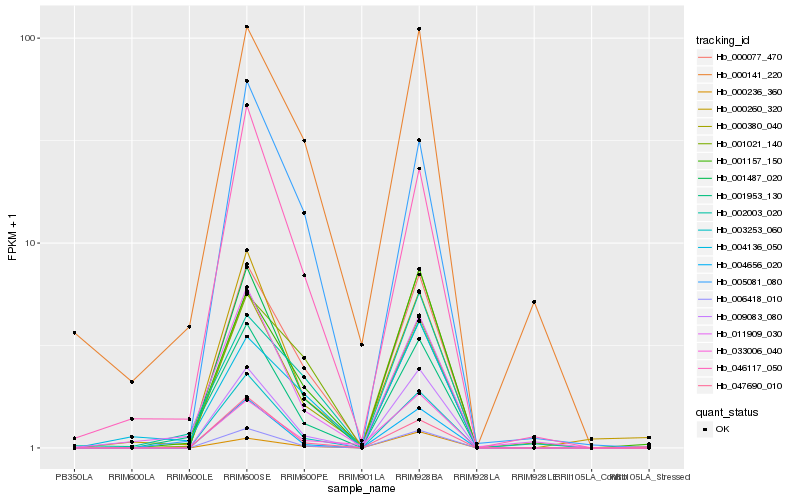
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_470 |
0.0 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 2 |
Hb_009083_080 |
0.0948792758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003253_060 |
0.1029647343 |
- |
- |
PREDICTED: tropinone reductase-like 1 [Vitis vinifera] |
| 4 |
Hb_001157_150 |
0.1147070777 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 5 |
Hb_001021_140 |
0.1150987122 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 6 |
Hb_005081_080 |
0.1210710599 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 7 |
Hb_006418_010 |
0.129367835 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 8 |
Hb_000380_040 |
0.1307527207 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 9 |
Hb_001487_020 |
0.1315598488 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001953_130 |
0.1332003188 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 11 |
Hb_004656_020 |
0.1370698403 |
- |
- |
EF hand domain-containing protein, variant [Neurospora crassa OR74A] |
| 12 |
Hb_033006_040 |
0.1404537114 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 13 |
Hb_046117_050 |
0.1472347161 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 14 |
Hb_047690_010 |
0.1503553578 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 15 |
Hb_000141_220 |
0.1532763597 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 16 |
Hb_002003_020 |
0.1533255099 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 17 |
Hb_004136_050 |
0.1542196641 |
- |
- |
- |
| 18 |
Hb_000260_320 |
0.1556000635 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 19 |
Hb_011909_030 |
0.1569396521 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |
| 20 |
Hb_000236_360 |
0.1584517576 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Prunus mume] |