| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
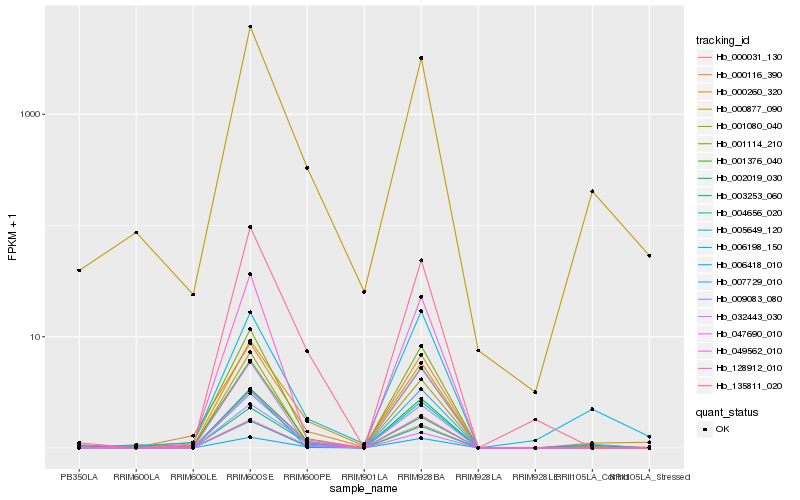
Hb_002019_030 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 2 |
Hb_006418_010 |
0.0955337785 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 3 |
Hb_004656_020 |
0.1110798719 |
- |
- |
EF hand domain-containing protein, variant [Neurospora crassa OR74A] |
| 4 |
Hb_003253_060 |
0.1192077001 |
- |
- |
PREDICTED: tropinone reductase-like 1 [Vitis vinifera] |
| 5 |
Hb_000877_090 |
0.1279035592 |
- |
- |
allergenic-related protein Pt2L4 [Manihot esculenta] |
| 6 |
Hb_001114_210 |
0.1293695603 |
- |
- |
PREDICTED: periaxin [Jatropha curcas] |
| 7 |
Hb_009083_080 |
0.1300612103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001376_040 |
0.1321462686 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 9 |
Hb_000031_130 |
0.1372185604 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 10 |
Hb_007729_010 |
0.1388228464 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_128912_010 |
0.1408537126 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 12 |
Hb_000260_320 |
0.1423036519 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 13 |
Hb_135811_020 |
0.1430092008 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 14 |
Hb_047690_010 |
0.1451951779 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 15 |
Hb_032443_030 |
0.1452752728 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 16 |
Hb_000116_390 |
0.1469376211 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 17 |
Hb_006198_150 |
0.1479901954 |
- |
- |
- |
| 18 |
Hb_005649_120 |
0.1521520227 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 19 |
Hb_049562_010 |
0.1532531412 |
- |
- |
hypothetical protein JCGZ_25442 [Jatropha curcas] |
| 20 |
Hb_001080_040 |
0.1534684048 |
- |
- |
cytochrome P450, putative [Ricinus communis] |