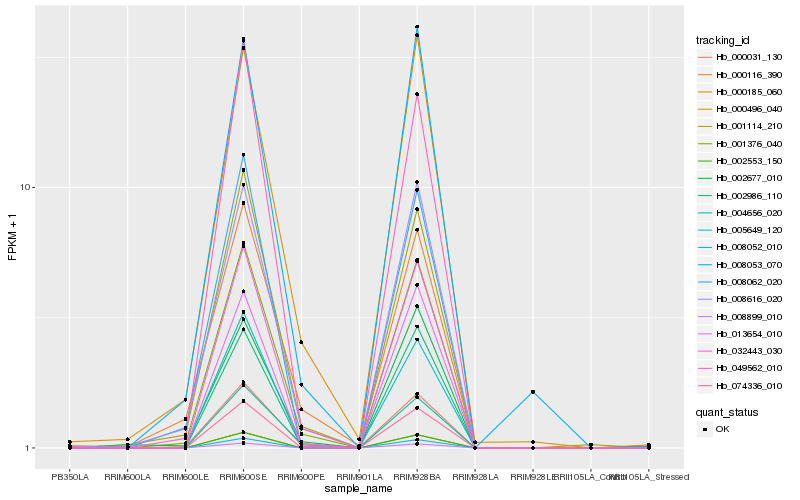
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001376_040 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 2 |
Hb_000031_130 |
0.0513427341 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 3 |
Hb_008616_020 |
0.0589011862 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001114_210 |
0.060827768 |
- |
- |
PREDICTED: periaxin [Jatropha curcas] |
| 5 |
Hb_032443_030 |
0.06451468 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 6 |
Hb_004656_020 |
0.0668177607 |
- |
- |
EF hand domain-containing protein, variant [Neurospora crassa OR74A] |
| 7 |
Hb_005649_120 |
0.0739439739 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002986_110 |
0.0821158289 |
- |
- |
PREDICTED: patatin-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000185_060 |
0.0837307744 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 10 |
Hb_049562_010 |
0.0853919584 |
- |
- |
hypothetical protein JCGZ_25442 [Jatropha curcas] |
| 11 |
Hb_002677_010 |
0.0908521615 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 12 |
Hb_008053_070 |
0.0911849559 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X4 [Jatropha curcas] |
| 13 |
Hb_008062_020 |
0.0929671442 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 14 |
Hb_013654_010 |
0.0936410392 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000116_390 |
0.0936881216 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 16 |
Hb_002553_150 |
0.0956470967 |
- |
- |
Gram-positive signal peptide protein, YSIRK family [Streptococcus pneumoniae TCH8431/19A] |
| 17 |
Hb_000496_040 |
0.0956527309 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008052_010 |
0.0956651076 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_074336_010 |
0.0956739766 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 20 |
Hb_008899_010 |
0.0957207706 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Nelumbo nucifera] |