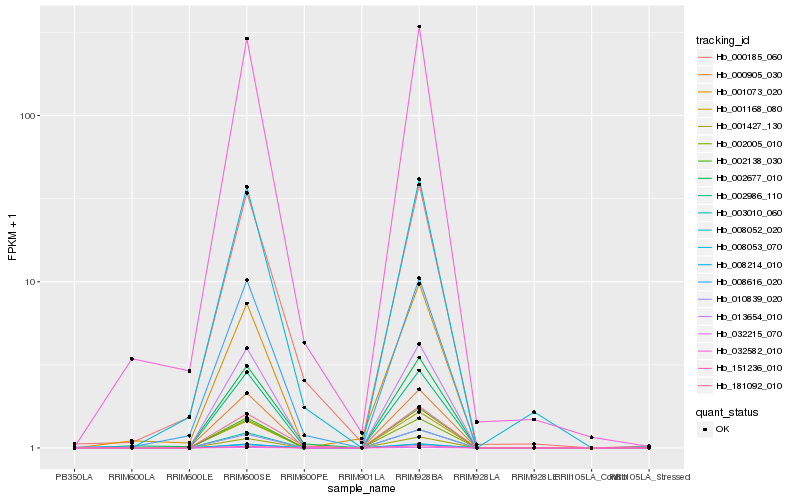
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032582_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 2 |
Hb_002677_010 |
0.0482022951 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 3 |
Hb_008616_020 |
0.0696514249 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_002986_110 |
0.0729432369 |
- |
- |
PREDICTED: patatin-like protein 1 [Jatropha curcas] |
| 5 |
Hb_013654_010 |
0.0732509908 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000905_030 |
0.0763633904 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 7 |
Hb_000185_060 |
0.0768443091 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008053_070 |
0.0799165036 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X4 [Jatropha curcas] |
| 9 |
Hb_001427_130 |
0.0839786532 |
- |
- |
PREDICTED: uncharacterized protein LOC101510272 [Cicer arietinum] |
| 10 |
Hb_032215_070 |
0.0839929832 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_003010_060 |
0.0845145445 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 12 |
Hb_008052_020 |
0.0857767004 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 13 |
Hb_008214_010 |
0.085883031 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 14 |
Hb_002005_010 |
0.0870268404 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39-like isoform X2 [Populus euphratica] |
| 15 |
Hb_001168_080 |
0.0877356065 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_181092_010 |
0.0879778823 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_010839_020 |
0.0910123337 |
- |
- |
- |
| 18 |
Hb_001073_020 |
0.0914015013 |
- |
- |
hypothetical protein CISIN_1g031854mg [Citrus sinensis] |
| 19 |
Hb_002138_030 |
0.0919372727 |
- |
- |
- |
| 20 |
Hb_151236_010 |
0.0934549127 |
- |
- |
hypothetical protein POPTR_0010s11560g [Populus trichocarpa] |