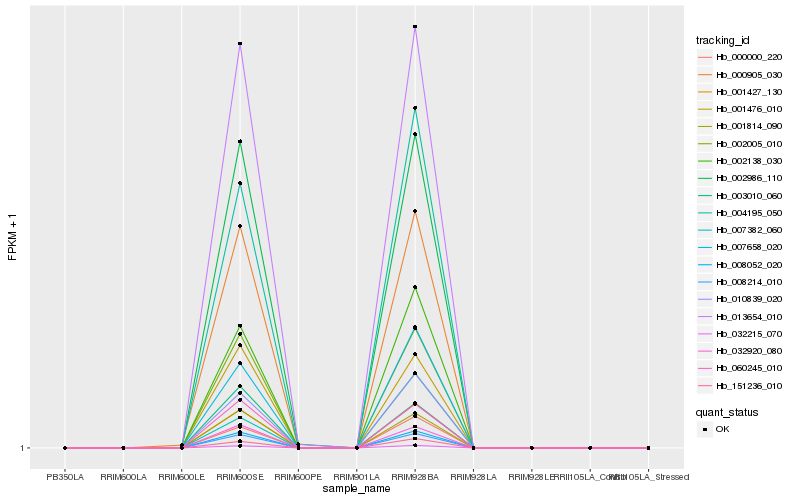
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101510272 [Cicer arietinum] |
| 2 |
Hb_032215_070 |
0.0028153856 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_003010_060 |
0.0086088327 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 4 |
Hb_008052_020 |
0.0185371505 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 5 |
Hb_008214_010 |
0.01905299 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 6 |
Hb_002005_010 |
0.0239100264 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39-like isoform X2 [Populus euphratica] |
| 7 |
Hb_010839_020 |
0.034244168 |
- |
- |
- |
| 8 |
Hb_002138_030 |
0.0365856179 |
- |
- |
- |
| 9 |
Hb_013654_010 |
0.0368777246 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_151236_010 |
0.040184968 |
- |
- |
hypothetical protein POPTR_0010s11560g [Populus trichocarpa] |
| 11 |
Hb_000905_030 |
0.0477163248 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 12 |
Hb_004195_050 |
0.0486419107 |
- |
- |
- |
| 13 |
Hb_007382_060 |
0.0486818482 |
- |
- |
- |
| 14 |
Hb_002986_110 |
0.0499131288 |
- |
- |
PREDICTED: patatin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000000_220 |
0.0522621494 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_007658_020 |
0.0525900805 |
- |
- |
Beta-amyrin synthase [Glycine soja] |
| 17 |
Hb_032920_080 |
0.0548175567 |
- |
- |
PREDICTED: uncharacterized protein LOC104602705 [Nelumbo nucifera] |
| 18 |
Hb_001814_090 |
0.0569811599 |
- |
- |
PREDICTED: uncharacterized protein LOC105636644 [Jatropha curcas] |
| 19 |
Hb_060245_010 |
0.0574011691 |
- |
- |
- |
| 20 |
Hb_001476_010 |
0.0587222999 |
- |
- |
hypothetical protein JCGZ_15669 [Jatropha curcas] |