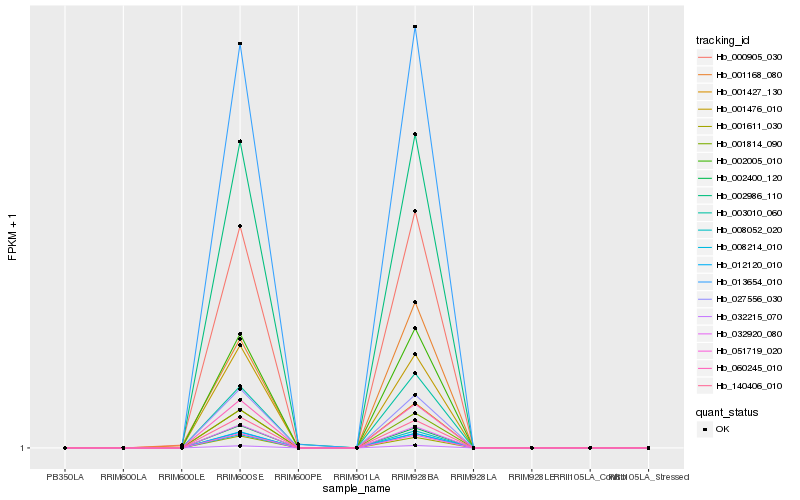
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000905_030 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 2 |
Hb_008052_020 |
0.0449666053 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 3 |
Hb_008214_010 |
0.0449968087 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 4 |
Hb_002005_010 |
0.0455669783 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39-like isoform X2 [Populus euphratica] |
| 5 |
Hb_032215_070 |
0.0468398658 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_001427_130 |
0.0477163248 |
- |
- |
PREDICTED: uncharacterized protein LOC101510272 [Cicer arietinum] |
| 7 |
Hb_003010_060 |
0.0512676229 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 8 |
Hb_013654_010 |
0.0547184492 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_001168_080 |
0.0576469011 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_032920_080 |
0.0592360699 |
- |
- |
PREDICTED: uncharacterized protein LOC104602705 [Nelumbo nucifera] |
| 11 |
Hb_001814_090 |
0.0606686597 |
- |
- |
PREDICTED: uncharacterized protein LOC105636644 [Jatropha curcas] |
| 12 |
Hb_060245_010 |
0.060951756 |
- |
- |
- |
| 13 |
Hb_001476_010 |
0.0618523697 |
- |
- |
hypothetical protein JCGZ_15669 [Jatropha curcas] |
| 14 |
Hb_002986_110 |
0.0620942389 |
- |
- |
PREDICTED: patatin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_012120_010 |
0.0621901728 |
- |
- |
- |
| 16 |
Hb_140406_010 |
0.0623455682 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 17 |
Hb_051719_020 |
0.0629696897 |
- |
- |
PREDICTED: toll/interleukin-1 receptor-like protein [Jatropha curcas] |
| 18 |
Hb_027556_030 |
0.0636498989 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like, partial [Gossypium raimondii] |
| 19 |
Hb_001611_030 |
0.0640300193 |
- |
- |
- |
| 20 |
Hb_002400_120 |
0.0642433134 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |