| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
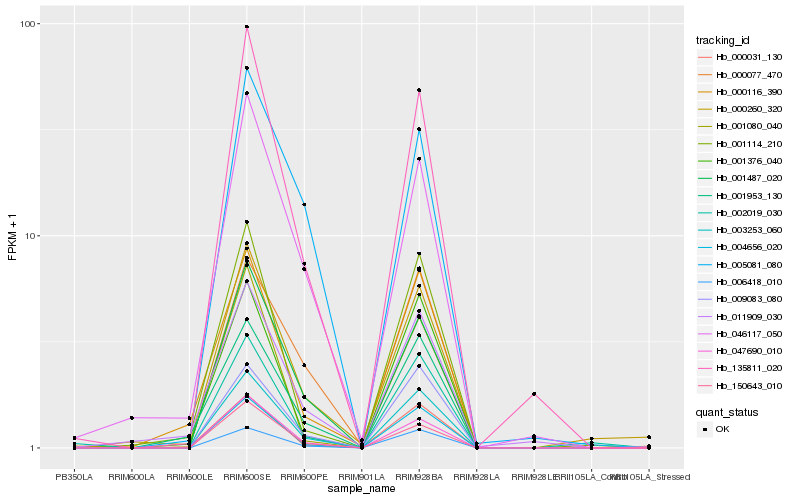
Hb_003253_060 |
0.0 |
- |
- |
PREDICTED: tropinone reductase-like 1 [Vitis vinifera] |
| 2 |
Hb_004656_020 |
0.0519611089 |
- |
- |
EF hand domain-containing protein, variant [Neurospora crassa OR74A] |
| 3 |
Hb_047690_010 |
0.0700036624 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 4 |
Hb_009083_080 |
0.0719281159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001487_020 |
0.0792447231 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_135811_020 |
0.0860904372 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 7 |
Hb_150643_010 |
0.0884590464 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 8 |
Hb_006418_010 |
0.0992063984 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 9 |
Hb_000077_470 |
0.1029647343 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 10 |
Hb_000031_130 |
0.1078665613 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 11 |
Hb_001376_040 |
0.1088526383 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 12 |
Hb_001953_130 |
0.1095048901 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 13 |
Hb_001114_210 |
0.1189203503 |
- |
- |
PREDICTED: periaxin [Jatropha curcas] |
| 14 |
Hb_002019_030 |
0.1192077001 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 15 |
Hb_000260_320 |
0.1200156346 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 16 |
Hb_000116_390 |
0.1227873975 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 17 |
Hb_046117_050 |
0.1228614806 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 18 |
Hb_001080_040 |
0.1272408774 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_005081_080 |
0.1347161071 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 20 |
Hb_011909_030 |
0.134780388 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |