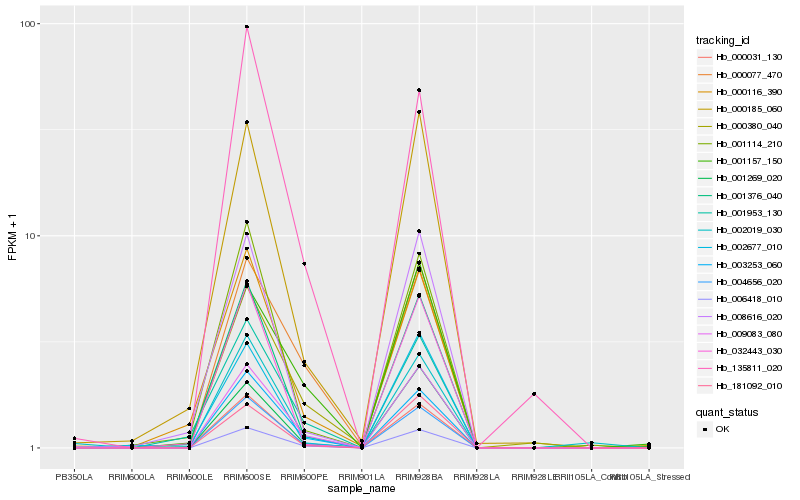
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006418_010 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 2 |
Hb_009083_080 |
0.084077087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004656_020 |
0.0955204368 |
- |
- |
EF hand domain-containing protein, variant [Neurospora crassa OR74A] |
| 4 |
Hb_002019_030 |
0.0955337785 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 5 |
Hb_003253_060 |
0.0992063984 |
- |
- |
PREDICTED: tropinone reductase-like 1 [Vitis vinifera] |
| 6 |
Hb_001376_040 |
0.1227308054 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 7 |
Hb_000185_060 |
0.1250234132 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 8 |
Hb_181092_010 |
0.1263641629 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000031_130 |
0.1291276241 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 10 |
Hb_000077_470 |
0.129367835 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 11 |
Hb_000380_040 |
0.1313596214 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 12 |
Hb_000116_390 |
0.1341111264 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 13 |
Hb_001953_130 |
0.1369108901 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 14 |
Hb_008616_020 |
0.1393507372 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_001269_020 |
0.140424123 |
- |
- |
PREDICTED: beta-Amyrin Synthase 2-like [Vitis vinifera] |
| 16 |
Hb_032443_030 |
0.1430991788 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 17 |
Hb_001157_150 |
0.1466059736 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 18 |
Hb_135811_020 |
0.1475154312 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 19 |
Hb_002677_010 |
0.1478002289 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 20 |
Hb_001114_210 |
0.1479209512 |
- |
- |
PREDICTED: periaxin [Jatropha curcas] |