| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
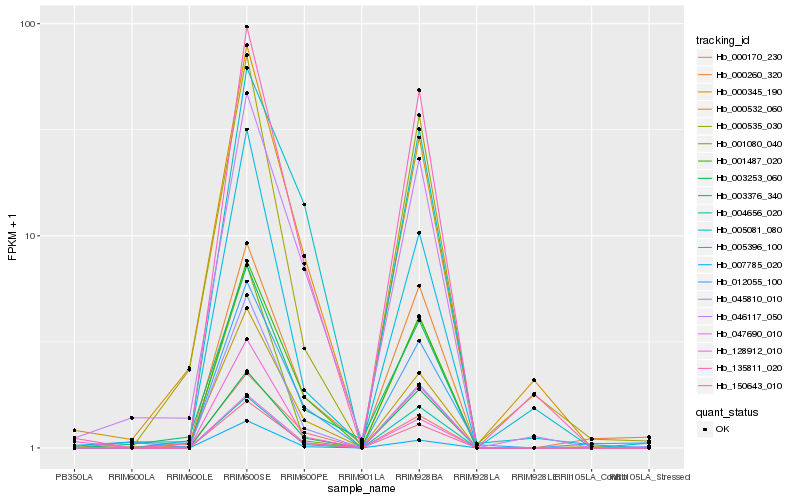
Hb_150643_010 |
0.0 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 2 |
Hb_047690_010 |
0.0186638892 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 3 |
Hb_001487_020 |
0.0413503805 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_135811_020 |
0.0631023869 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 5 |
Hb_003253_060 |
0.0884590464 |
- |
- |
PREDICTED: tropinone reductase-like 1 [Vitis vinifera] |
| 6 |
Hb_046117_050 |
0.1031662222 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 7 |
Hb_007785_020 |
0.1036238328 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_128912_010 |
0.105570099 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 9 |
Hb_000532_060 |
0.1060892046 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 10 |
Hb_001080_040 |
0.1139183049 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_003376_340 |
0.1171072397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_045810_010 |
0.1176778251 |
- |
- |
PREDICTED: laccase-12-like [Jatropha curcas] |
| 13 |
Hb_000345_190 |
0.1196117091 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 14 |
Hb_004656_020 |
0.1200820264 |
- |
- |
EF hand domain-containing protein, variant [Neurospora crassa OR74A] |
| 15 |
Hb_005081_080 |
0.1243510558 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 16 |
Hb_000170_230 |
0.1254446815 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 17 |
Hb_012055_100 |
0.1256242795 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 18 |
Hb_000260_320 |
0.1293743618 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 19 |
Hb_000535_030 |
0.1312671022 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 20 |
Hb_005396_100 |
0.1345914984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |