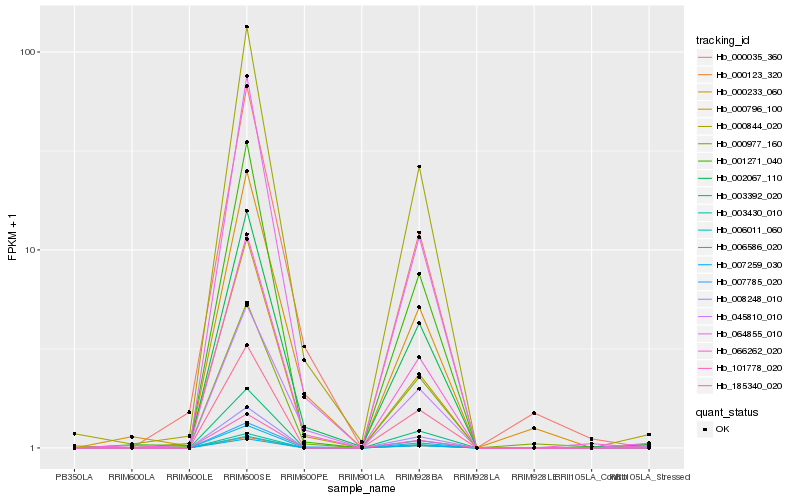
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002067_110 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 2 |
Hb_000844_020 |
0.0436926486 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 3 |
Hb_007785_020 |
0.0690324526 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_066262_020 |
0.069206726 |
- |
- |
hypothetical protein JCGZ_14803 [Jatropha curcas] |
| 5 |
Hb_064855_010 |
0.0783619663 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_045810_010 |
0.0799051748 |
- |
- |
PREDICTED: laccase-12-like [Jatropha curcas] |
| 7 |
Hb_001271_040 |
0.0818569837 |
- |
- |
hypothetical protein POPTR_0019s039903g, partial [Populus trichocarpa] |
| 8 |
Hb_000977_160 |
0.0920426852 |
- |
- |
Serine-threonine protein kinase, plant-type, putative [Theobroma cacao] |
| 9 |
Hb_000796_100 |
0.0981930323 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 10 |
Hb_006011_060 |
0.0987707009 |
- |
- |
- |
| 11 |
Hb_003392_020 |
0.0987759258 |
- |
- |
PREDICTED: uncharacterized protein LOC105650609 [Jatropha curcas] |
| 12 |
Hb_008248_010 |
0.0990456082 |
- |
- |
- |
| 13 |
Hb_003430_010 |
0.0990646712 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 33 precursor [Populus trichocarpa] |
| 14 |
Hb_000123_320 |
0.0991911088 |
- |
- |
PREDICTED: MADS-box protein FLOWERING LOCUS C-like [Jatropha curcas] |
| 15 |
Hb_185340_020 |
0.0999278621 |
- |
- |
- |
| 16 |
Hb_007259_030 |
0.0999709889 |
- |
- |
- |
| 17 |
Hb_006586_020 |
0.1001291034 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_000035_360 |
0.1008675378 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 19 |
Hb_101778_020 |
0.1009182527 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000233_060 |
0.101380223 |
- |
- |
hypothetical protein JCGZ_14793 [Jatropha curcas] |