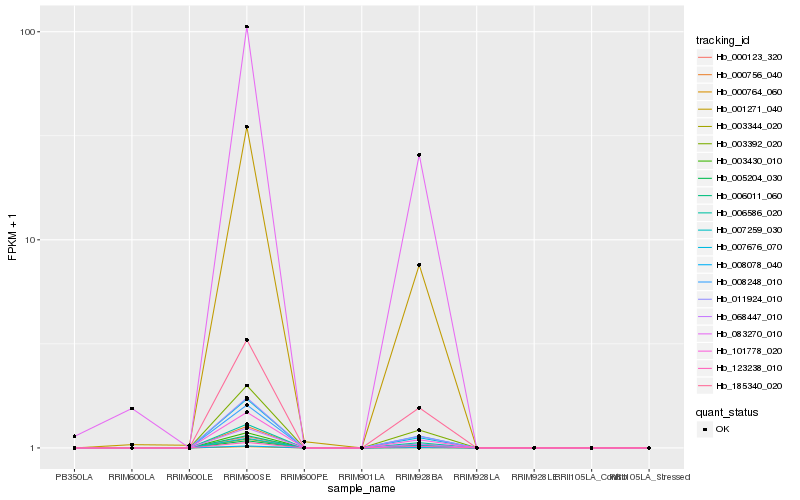
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003430_010 |
0.0 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 33 precursor [Populus trichocarpa] |
| 2 |
Hb_000123_320 |
0.0015021306 |
- |
- |
PREDICTED: MADS-box protein FLOWERING LOCUS C-like [Jatropha curcas] |
| 3 |
Hb_003392_020 |
0.0065709833 |
- |
- |
PREDICTED: uncharacterized protein LOC105650609 [Jatropha curcas] |
| 4 |
Hb_006011_060 |
0.0072547301 |
- |
- |
- |
| 5 |
Hb_006586_020 |
0.0088487106 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_101778_020 |
0.0131401376 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008248_010 |
0.0150970873 |
- |
- |
- |
| 8 |
Hb_011924_010 |
0.020032177 |
- |
- |
hypothetical protein, partial [Phaeodactylibacter xiamenensis] |
| 9 |
Hb_185340_020 |
0.0229173871 |
- |
- |
- |
| 10 |
Hb_007259_030 |
0.0232002845 |
- |
- |
- |
| 11 |
Hb_068447_010 |
0.0327306206 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_007676_070 |
0.0364286702 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, endoplasmic reticulum isozyme 1-like [Populus euphratica] |
| 13 |
Hb_000764_060 |
0.0378935945 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |
| 14 |
Hb_000756_040 |
0.0419651837 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_008078_040 |
0.0433302936 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 16 |
Hb_001271_040 |
0.0435438879 |
- |
- |
hypothetical protein POPTR_0019s039903g, partial [Populus trichocarpa] |
| 17 |
Hb_005204_030 |
0.0475141132 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |
| 18 |
Hb_123238_010 |
0.0540517191 |
- |
- |
hypothetical protein JCGZ_16598 [Jatropha curcas] |
| 19 |
Hb_083270_010 |
0.0542246942 |
- |
- |
hypothetical protein ARALYDRAFT_896220 [Arabidopsis lyrata subsp. lyrata] |
| 20 |
Hb_003344_020 |
0.0560704506 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103501197 [Cucumis melo] |