| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
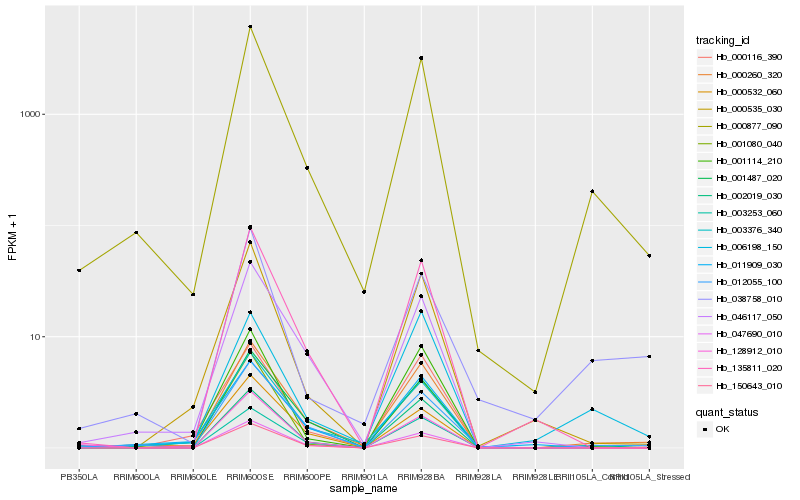
Hb_000877_090 |
0.0 |
- |
- |
allergenic-related protein Pt2L4 [Manihot esculenta] |
| 2 |
Hb_000260_320 |
0.0857824055 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 3 |
Hb_012055_100 |
0.1089840052 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 4 |
Hb_002019_030 |
0.1279035592 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 5 |
Hb_003376_340 |
0.1477785456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000532_060 |
0.148728726 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |
| 7 |
Hb_046117_050 |
0.1489159247 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 8 |
Hb_135811_020 |
0.1501965011 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 9 |
Hb_001114_210 |
0.1502611091 |
- |
- |
PREDICTED: periaxin [Jatropha curcas] |
| 10 |
Hb_047690_010 |
0.1515167386 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 11 |
Hb_011909_030 |
0.1535919287 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |
| 12 |
Hb_150643_010 |
0.154912197 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000535_030 |
0.155061164 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 14 |
Hb_006198_150 |
0.1593226125 |
- |
- |
- |
| 15 |
Hb_003253_060 |
0.1603591274 |
- |
- |
PREDICTED: tropinone reductase-like 1 [Vitis vinifera] |
| 16 |
Hb_038758_010 |
0.1605152987 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_128912_010 |
0.1613120061 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 18 |
Hb_000116_390 |
0.1630395905 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 19 |
Hb_001487_020 |
0.1635201999 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001080_040 |
0.1639073271 |
- |
- |
cytochrome P450, putative [Ricinus communis] |