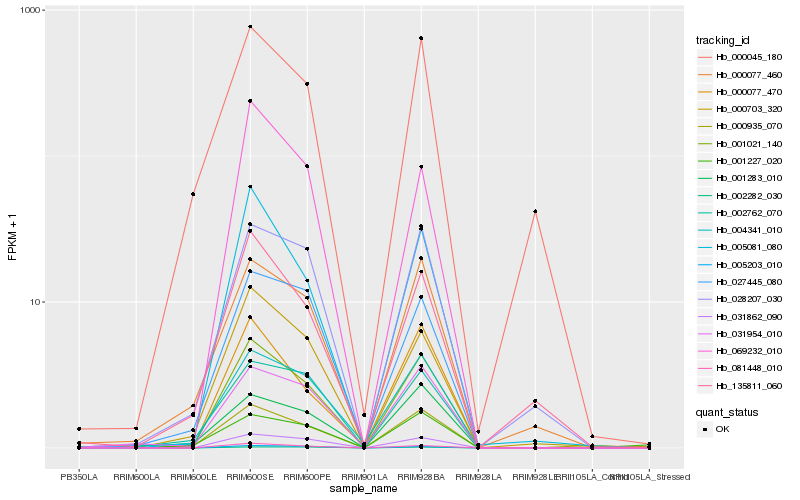
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001021_140 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 2 |
Hb_081448_010 |
0.0833122402 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 3 |
Hb_031862_090 |
0.0951695953 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 4 |
Hb_005081_080 |
0.1049281146 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 5 |
Hb_000703_320 |
0.114978037 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 6 |
Hb_000077_470 |
0.1150987122 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 7 |
Hb_004341_010 |
0.1151011047 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 8 |
Hb_000935_070 |
0.1163110939 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_031954_010 |
0.1216219111 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_005203_010 |
0.1275191668 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 11 |
Hb_028207_030 |
0.12769423 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 12 |
Hb_000077_460 |
0.1325600545 |
- |
- |
- |
| 13 |
Hb_027445_080 |
0.1387919399 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 14 |
Hb_069232_010 |
0.1400159071 |
- |
- |
PREDICTED: 60S ribosomal protein L37-3-like [Vitis vinifera] |
| 15 |
Hb_002762_070 |
0.1415690795 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 16 |
Hb_135811_060 |
0.143353126 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 17 |
Hb_001283_010 |
0.1500527953 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 18 |
Hb_000045_180 |
0.1550592379 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 19 |
Hb_001227_020 |
0.1563716948 |
- |
- |
- |
| 20 |
Hb_002282_030 |
0.1573714054 |
- |
- |
- |