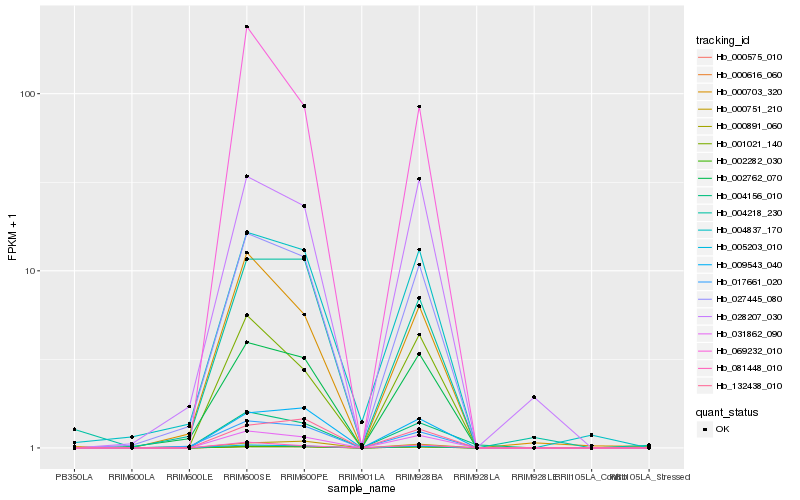
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005203_010 |
0.0 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 2 |
Hb_031862_090 |
0.0475944494 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 3 |
Hb_027445_080 |
0.0777278441 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 4 |
Hb_002282_030 |
0.0792153768 |
- |
- |
- |
| 5 |
Hb_000891_060 |
0.0835305172 |
- |
- |
hypothetical protein JCGZ_11234 [Jatropha curcas] |
| 6 |
Hb_000616_060 |
0.0863968886 |
- |
- |
- |
| 7 |
Hb_000575_010 |
0.0873324686 |
- |
- |
PREDICTED: uncharacterized protein LOC103454030 [Malus domestica] |
| 8 |
Hb_081448_010 |
0.0982211759 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 9 |
Hb_009543_040 |
0.1086286895 |
- |
- |
- |
| 10 |
Hb_017661_020 |
0.1115202482 |
- |
- |
hypothetical protein JCGZ_11176 [Jatropha curcas] |
| 11 |
Hb_002762_070 |
0.114878455 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 12 |
Hb_069232_010 |
0.1190432969 |
- |
- |
PREDICTED: 60S ribosomal protein L37-3-like [Vitis vinifera] |
| 13 |
Hb_000703_320 |
0.1227347105 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 14 |
Hb_001021_140 |
0.1275191668 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 15 |
Hb_132438_010 |
0.1312740486 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 16 |
Hb_004837_170 |
0.1319249189 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 17 |
Hb_004156_010 |
0.1349655731 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 18 |
Hb_000751_210 |
0.1366570934 |
- |
- |
PREDICTED: petal death protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_004218_230 |
0.1383236159 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 20 |
Hb_028207_030 |
0.1395287184 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |