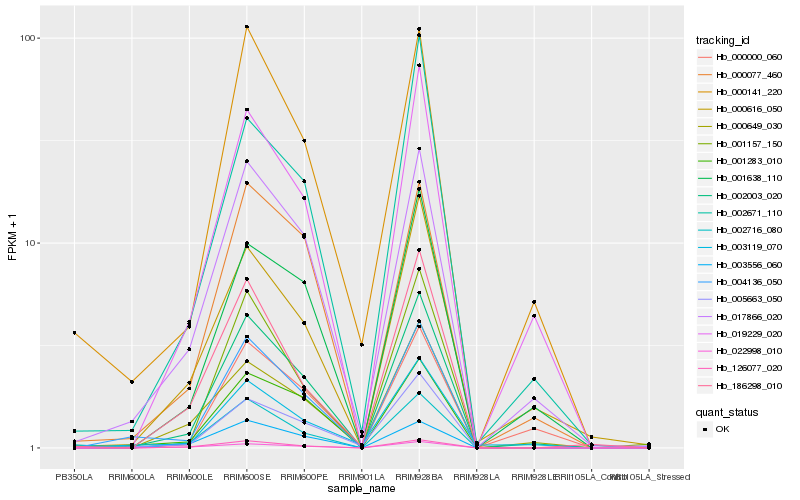
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002003_020 |
0.0 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 2 |
Hb_017866_020 |
0.0847938105 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 3 |
Hb_019229_020 |
0.095632685 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_004136_050 |
0.1049808611 |
- |
- |
- |
| 5 |
Hb_000077_460 |
0.1052544578 |
- |
- |
- |
| 6 |
Hb_001638_110 |
0.1075118741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001283_010 |
0.1079508694 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 8 |
Hb_002716_080 |
0.110407556 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 9 |
Hb_126077_020 |
0.1134247645 |
- |
- |
Kinase family protein with leucine-rich repeat domain [Theobroma cacao] |
| 10 |
Hb_000141_220 |
0.1156516309 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 11 |
Hb_000616_050 |
0.1191533364 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 12 |
Hb_001157_150 |
0.1193036344 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 13 |
Hb_000649_030 |
0.1225618856 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 14 |
Hb_002671_110 |
0.1230505532 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 15 |
Hb_000000_060 |
0.1257057604 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 16 |
Hb_186298_010 |
0.1267274822 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_005663_050 |
0.1275942321 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_022998_010 |
0.1296386105 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_003119_070 |
0.1298531402 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_003556_060 |
0.1315773748 |
- |
- |
- |