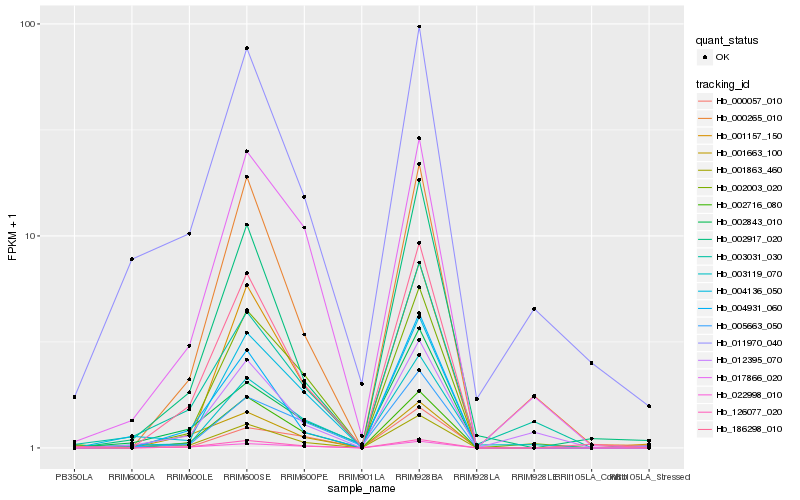
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_460 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 2 |
Hb_004136_050 |
0.1005576973 |
- |
- |
- |
| 3 |
Hb_186298_010 |
0.1211197371 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_126077_020 |
0.1300926263 |
- |
- |
Kinase family protein with leucine-rich repeat domain [Theobroma cacao] |
| 5 |
Hb_002917_020 |
0.1372943881 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 6 |
Hb_002843_010 |
0.1389295329 |
- |
- |
hypothetical protein EUGRSUZ_H04984, partial [Eucalyptus grandis] |
| 7 |
Hb_004931_060 |
0.1400350841 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 8 |
Hb_002003_020 |
0.1400985398 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 9 |
Hb_011970_040 |
0.1453569723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_022998_010 |
0.1462937568 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_003031_030 |
0.1532284836 |
- |
- |
hypothetical protein JCGZ_21288 [Jatropha curcas] |
| 12 |
Hb_001157_150 |
0.1566597205 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 13 |
Hb_002716_080 |
0.1573270843 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 14 |
Hb_012395_070 |
0.1577201471 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL6 [Jatropha curcas] |
| 15 |
Hb_000265_010 |
0.1581802105 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_017866_020 |
0.1594130458 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 17 |
Hb_005663_050 |
0.1595403374 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_001663_100 |
0.1631422385 |
- |
- |
- |
| 19 |
Hb_000057_010 |
0.1632136737 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 20 |
Hb_003119_070 |
0.1642949484 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |