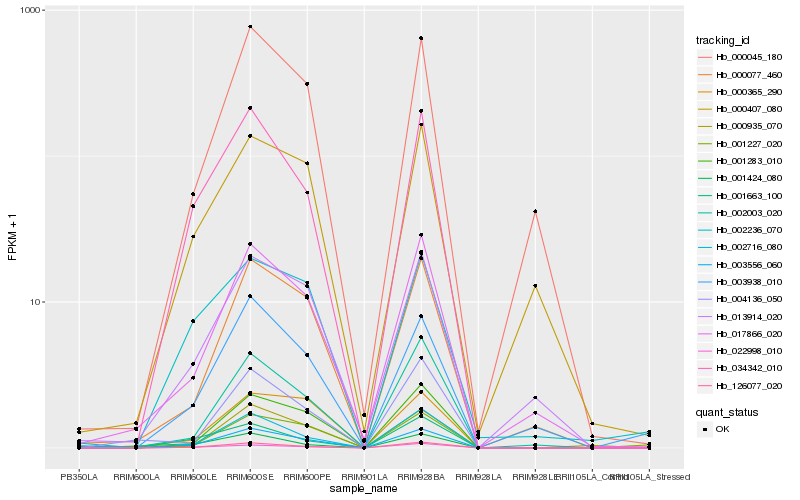
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003556_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_126077_020 |
0.0737141308 |
- |
- |
Kinase family protein with leucine-rich repeat domain [Theobroma cacao] |
| 3 |
Hb_034342_010 |
0.0792767355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_022998_010 |
0.1017467434 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_001424_080 |
0.1085930344 |
- |
- |
PREDICTED: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 6 |
Hb_017866_020 |
0.1163050152 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 7 |
Hb_000077_460 |
0.1174885138 |
- |
- |
- |
| 8 |
Hb_001283_010 |
0.1222986884 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 9 |
Hb_000045_180 |
0.1227866859 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 10 |
Hb_002003_020 |
0.1315773748 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 11 |
Hb_013914_020 |
0.134759491 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 12 |
Hb_002716_080 |
0.1362293635 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 13 |
Hb_003938_010 |
0.1370363747 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor 1B, putative [Ricinus communis] |
| 14 |
Hb_001663_100 |
0.1379854199 |
- |
- |
- |
| 15 |
Hb_000935_070 |
0.1409031222 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000365_290 |
0.1446853825 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002236_070 |
0.1486621647 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 18 |
Hb_001227_020 |
0.1490699755 |
- |
- |
- |
| 19 |
Hb_000407_080 |
0.1510264689 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 20 |
Hb_004136_050 |
0.1551560938 |
- |
- |
- |