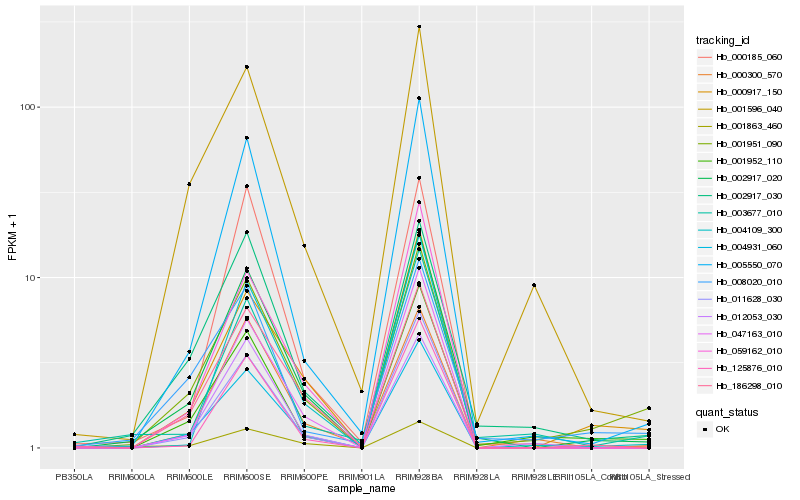
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002917_020 |
0.0 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 2 |
Hb_004931_060 |
0.0880475773 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 3 |
Hb_186298_010 |
0.1036726407 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_005550_070 |
0.1061348065 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 5 |
Hb_011628_030 |
0.1077386516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_047163_010 |
0.1114701034 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 7 |
Hb_001951_090 |
0.1154660009 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 8 |
Hb_002917_030 |
0.1186104678 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Setaria italica] |
| 9 |
Hb_000917_150 |
0.1192556016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_059162_010 |
0.1288715264 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_001952_110 |
0.1289379824 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_004109_300 |
0.1299882125 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 13 |
Hb_125876_010 |
0.1347518575 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001596_040 |
0.1351823159 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase 1-like [Populus euphratica] |
| 15 |
Hb_000185_060 |
0.136066662 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000300_570 |
0.136191213 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 17 |
Hb_001863_460 |
0.1372943881 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 18 |
Hb_003677_010 |
0.1400690173 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 19 |
Hb_012053_030 |
0.1417858036 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 20 |
Hb_008020_010 |
0.1428768541 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like [Jatropha curcas] |