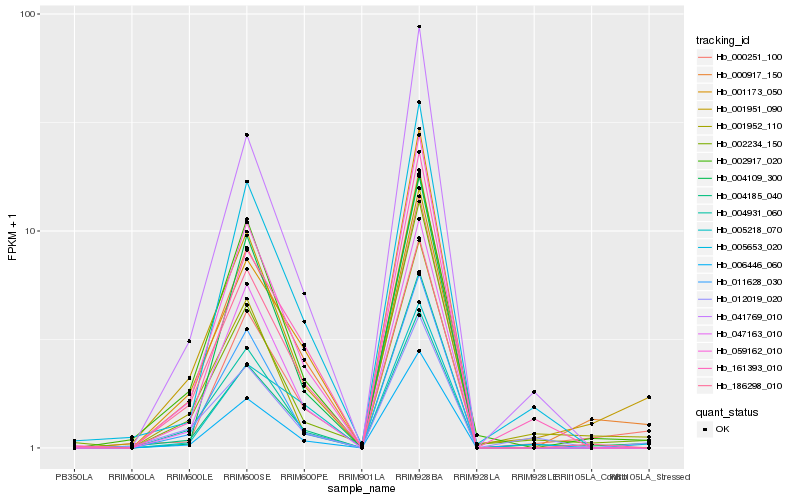
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000917_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002917_020 |
0.1192556016 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 3 |
Hb_059162_010 |
0.1193354511 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_047163_010 |
0.1237786007 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 5 |
Hb_001951_090 |
0.1251139559 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_005218_070 |
0.1357237887 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 7 |
Hb_004931_060 |
0.1370412358 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 8 |
Hb_011628_030 |
0.1383311865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001952_110 |
0.1391503397 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_041769_010 |
0.1395360523 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 11 |
Hb_186298_010 |
0.1408954481 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_004109_300 |
0.1420459592 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 13 |
Hb_000251_100 |
0.1433488393 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_161393_010 |
0.1443477205 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_005653_020 |
0.1456357807 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 16 |
Hb_002234_150 |
0.1465200167 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 17 |
Hb_001173_050 |
0.1491389451 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 18 |
Hb_006446_060 |
0.1526275906 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 19 |
Hb_004185_040 |
0.1539987941 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 20 |
Hb_012019_020 |
0.1552259879 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |