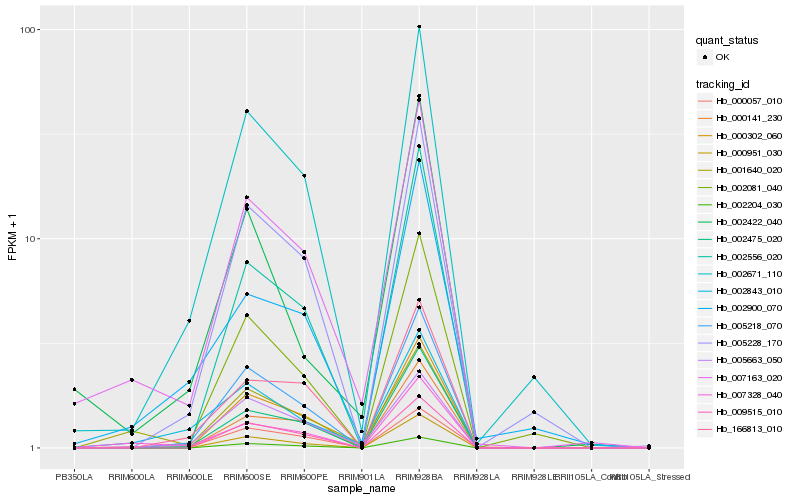
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007163_020 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_007328_040 |
0.0978279016 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Nelumbo nucifera] |
| 3 |
Hb_002671_110 |
0.1267159119 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 4 |
Hb_001640_020 |
0.126977409 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 5 |
Hb_000057_010 |
0.1333396656 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 6 |
Hb_002843_010 |
0.1340506206 |
- |
- |
hypothetical protein EUGRSUZ_H04984, partial [Eucalyptus grandis] |
| 7 |
Hb_005218_070 |
0.1373819287 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 8 |
Hb_002475_020 |
0.1410040246 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 9 |
Hb_002900_070 |
0.1463792827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000302_060 |
0.1487791738 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_005228_170 |
0.1497357635 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 12 |
Hb_005663_050 |
0.1503245657 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_002204_030 |
0.1521956263 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 14 |
Hb_166813_010 |
0.1556295485 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 15 |
Hb_002556_020 |
0.1559239437 |
- |
- |
hypothetical protein JCGZ_12588 [Jatropha curcas] |
| 16 |
Hb_009515_010 |
0.1564430732 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 17 |
Hb_000951_030 |
0.1579801721 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_000141_230 |
0.1580901512 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Camelina sativa] |
| 19 |
Hb_002081_040 |
0.1610291003 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 20 |
Hb_002422_040 |
0.1628529188 |
- |
- |
hypothetical protein POPTR_0016s02350g [Populus trichocarpa] |