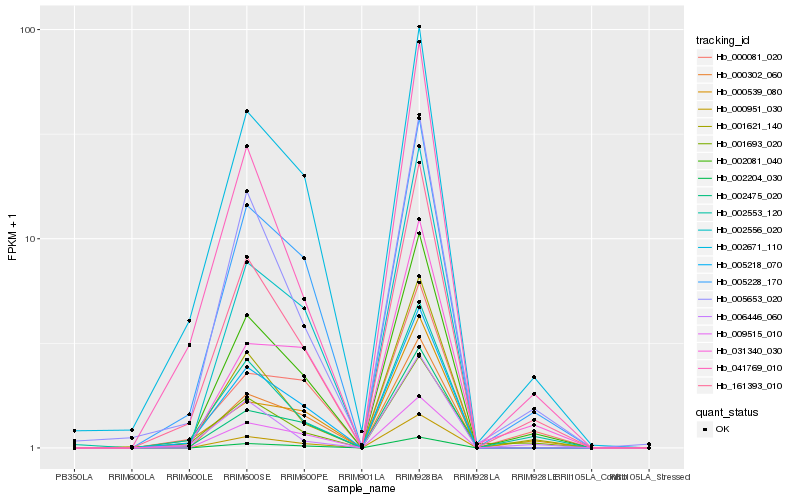
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002081_040 |
0.0 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 2 |
Hb_161393_010 |
0.048685584 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_005228_170 |
0.0609494172 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 4 |
Hb_001621_140 |
0.0863294588 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 5 |
Hb_005653_020 |
0.0878982482 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 6 |
Hb_005218_070 |
0.0898704068 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 7 |
Hb_000951_030 |
0.092874325 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_002671_110 |
0.0949650012 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 9 |
Hb_002204_030 |
0.0989689139 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 10 |
Hb_002553_120 |
0.0991686101 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B [Jatropha curcas] |
| 11 |
Hb_000302_060 |
0.0994290249 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_002475_020 |
0.1008094223 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 13 |
Hb_002556_020 |
0.1050872378 |
- |
- |
hypothetical protein JCGZ_12588 [Jatropha curcas] |
| 14 |
Hb_041769_010 |
0.1114734732 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 15 |
Hb_006446_060 |
0.1115514721 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 16 |
Hb_009515_010 |
0.1131754199 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 17 |
Hb_000081_020 |
0.1150219172 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 1.1-like [Camelina sativa] |
| 18 |
Hb_000539_080 |
0.1155215371 |
- |
- |
hypothetical protein POPTR_0039s00260g, partial [Populus trichocarpa] |
| 19 |
Hb_001693_020 |
0.1228140463 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_031340_030 |
0.1251698034 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Jatropha curcas] |