| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
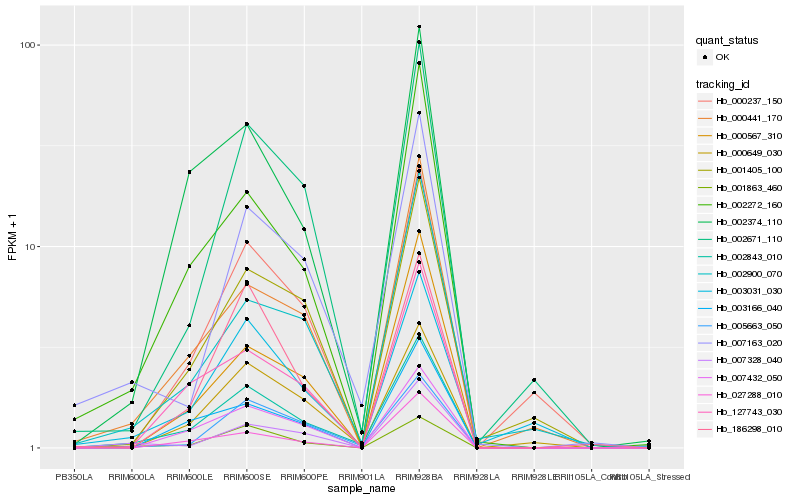
Hb_002843_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_H04984, partial [Eucalyptus grandis] |
| 2 |
Hb_002272_160 |
0.1228550207 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_002374_110 |
0.1283759808 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 4 |
Hb_007432_050 |
0.1290997582 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |
| 5 |
Hb_127743_030 |
0.1312334715 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_007163_020 |
0.1340506206 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_002671_110 |
0.1367663838 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 8 |
Hb_001863_460 |
0.1389295329 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 9 |
Hb_000441_170 |
0.1407148021 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 10 |
Hb_000649_030 |
0.1434807185 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 11 |
Hb_003166_040 |
0.1437488405 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 12 |
Hb_003031_030 |
0.1463982173 |
- |
- |
hypothetical protein JCGZ_21288 [Jatropha curcas] |
| 13 |
Hb_001405_100 |
0.1492366182 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 14 |
Hb_007328_040 |
0.1493004389 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Nelumbo nucifera] |
| 15 |
Hb_000237_150 |
0.1500825328 |
- |
- |
Epoxide hydrolase 2 [Morus notabilis] |
| 16 |
Hb_027288_010 |
0.1513155012 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 17 |
Hb_002900_070 |
0.1520719816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_186298_010 |
0.1539150104 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_005663_050 |
0.1546985971 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_000567_310 |
0.1566039073 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |