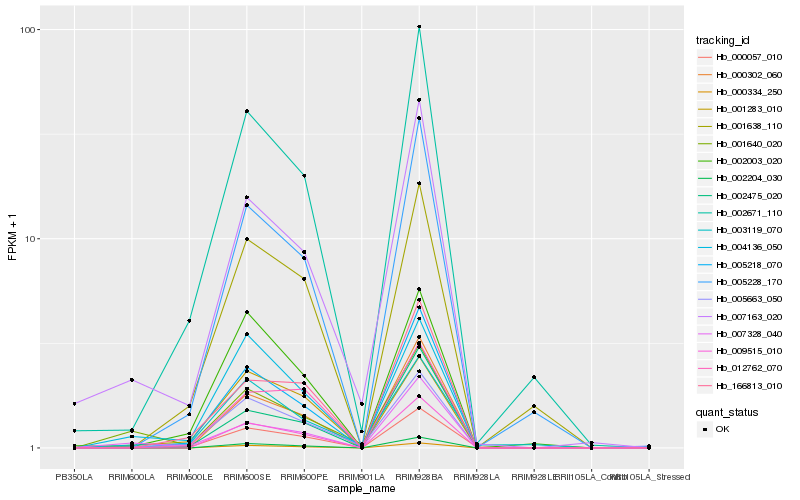
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000057_010 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 2 |
Hb_005218_070 |
0.1241781019 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 3 |
Hb_002671_110 |
0.1268460421 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 4 |
Hb_009515_010 |
0.132852109 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 5 |
Hb_007163_020 |
0.1333396656 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_004136_050 |
0.1342632497 |
- |
- |
- |
| 7 |
Hb_000334_250 |
0.1345494763 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 8 |
Hb_005228_170 |
0.1346628211 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 9 |
Hb_005663_050 |
0.1353367239 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_002204_030 |
0.1389241824 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 11 |
Hb_007328_040 |
0.1413519653 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Nelumbo nucifera] |
| 12 |
Hb_003119_070 |
0.1428230632 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 13 |
Hb_000302_060 |
0.1433228927 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001283_010 |
0.1445391058 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 15 |
Hb_166813_010 |
0.1453617336 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 16 |
Hb_012762_070 |
0.1503299213 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 17 |
Hb_002003_020 |
0.1504174507 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 18 |
Hb_001640_020 |
0.1507566512 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 19 |
Hb_001638_110 |
0.1538254806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002475_020 |
0.1539583614 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |