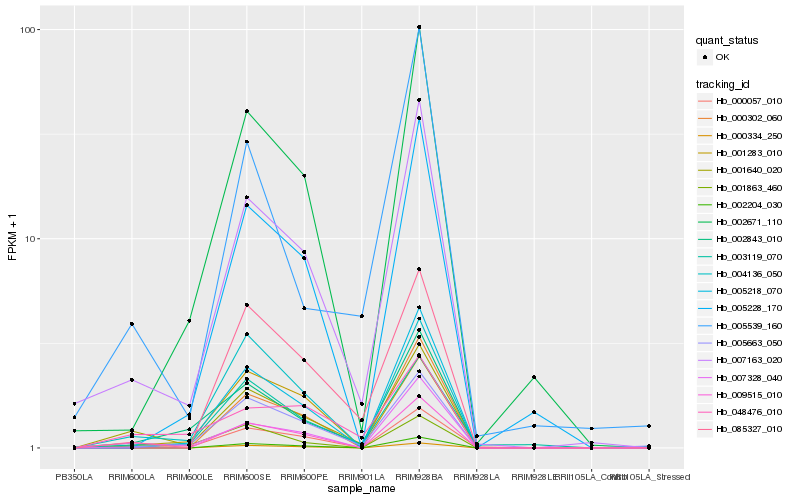
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001640_020 |
0.0 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 2 |
Hb_007163_020 |
0.126977409 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_007328_040 |
0.133217538 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Nelumbo nucifera] |
| 4 |
Hb_000057_010 |
0.1507566512 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 5 |
Hb_004136_050 |
0.1593286351 |
- |
- |
- |
| 6 |
Hb_005663_050 |
0.1613102233 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_002843_010 |
0.1651693723 |
- |
- |
hypothetical protein EUGRSUZ_H04984, partial [Eucalyptus grandis] |
| 8 |
Hb_001863_460 |
0.1691196397 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 9 |
Hb_085327_010 |
0.1700234275 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 10 |
Hb_048476_010 |
0.1723471448 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_003119_070 |
0.1799412862 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_002671_110 |
0.1844105133 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 13 |
Hb_005218_070 |
0.1866695101 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 14 |
Hb_009515_010 |
0.1952040442 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 15 |
Hb_002204_030 |
0.1954038747 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 16 |
Hb_005228_170 |
0.1971733663 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 17 |
Hb_000334_250 |
0.1974024583 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 18 |
Hb_000302_060 |
0.1991328955 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_005539_160 |
0.2005572268 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_001283_010 |
0.2043606765 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |