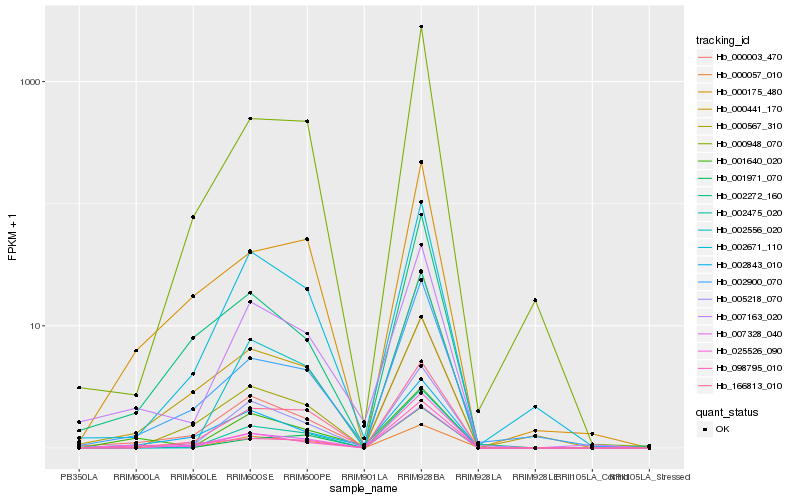
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007328_040 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Nelumbo nucifera] |
| 2 |
Hb_007163_020 |
0.0978279016 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_001640_020 |
0.133217538 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 4 |
Hb_002900_070 |
0.1336256605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000441_170 |
0.1355893539 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 6 |
Hb_002475_020 |
0.137651325 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 7 |
Hb_098795_010 |
0.1379786766 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase-like [Jatropha curcas] |
| 8 |
Hb_001971_070 |
0.1403805891 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_000057_010 |
0.1413519653 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 10 |
Hb_000175_480 |
0.1418587275 |
- |
- |
hypothetical protein JCGZ_06165 [Jatropha curcas] |
| 11 |
Hb_000003_470 |
0.1419557308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000948_070 |
0.147847485 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 13 |
Hb_002671_110 |
0.1479497001 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 14 |
Hb_005218_070 |
0.1485886991 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 15 |
Hb_002843_010 |
0.1493004389 |
- |
- |
hypothetical protein EUGRSUZ_H04984, partial [Eucalyptus grandis] |
| 16 |
Hb_166813_010 |
0.1515695463 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 17 |
Hb_002272_160 |
0.1530407932 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000567_310 |
0.1554839105 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 19 |
Hb_002556_020 |
0.1560386345 |
- |
- |
hypothetical protein JCGZ_12588 [Jatropha curcas] |
| 20 |
Hb_025526_090 |
0.1568918094 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |