| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
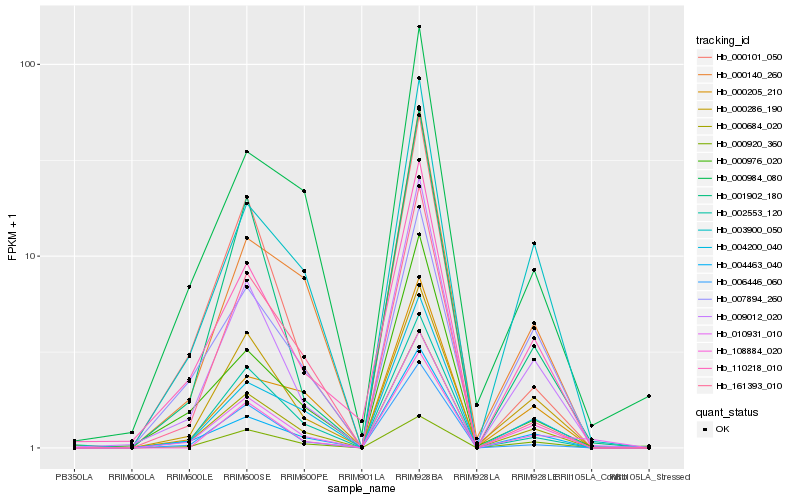
Hb_000684_020 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_110218_010 |
0.0927353722 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 3 |
Hb_004200_040 |
0.0935135425 |
- |
- |
laccase, putative [Ricinus communis] |
| 4 |
Hb_009012_020 |
0.0941569502 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 5 |
Hb_004463_040 |
0.0945670332 |
- |
- |
- |
| 6 |
Hb_002553_120 |
0.0962066039 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B [Jatropha curcas] |
| 7 |
Hb_003900_050 |
0.102719072 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 8 |
Hb_000140_260 |
0.1201825378 |
- |
- |
PREDICTED: uncharacterized protein LOC105642920 [Jatropha curcas] |
| 9 |
Hb_000286_190 |
0.1213973135 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 10 |
Hb_000205_210 |
0.1276934632 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_000920_360 |
0.1287238958 |
- |
- |
PREDICTED: (R,S)-reticuline 7-O-methyltransferase-like [Jatropha curcas] |
| 12 |
Hb_006446_060 |
0.1303297859 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 13 |
Hb_007894_260 |
0.131825005 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 14 |
Hb_161393_010 |
0.1325177883 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_010931_010 |
0.1348824013 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 16 |
Hb_001902_180 |
0.1356666434 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 17 |
Hb_000976_020 |
0.1362437623 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 18 |
Hb_108884_020 |
0.1365654258 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 19 |
Hb_000984_080 |
0.1382782455 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 20 |
Hb_000101_050 |
0.1389366578 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |