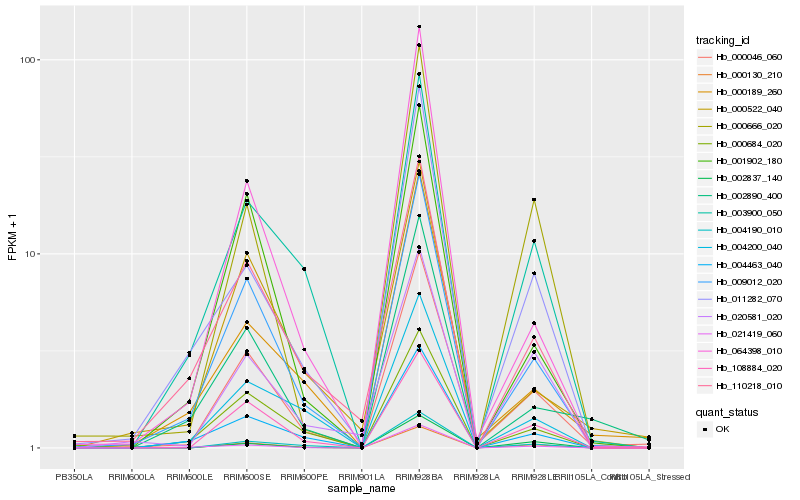
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_060 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_009012_020 |
0.0891684757 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 3 |
Hb_108884_020 |
0.1092136964 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 4 |
Hb_004190_010 |
0.1197421455 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000130_210 |
0.1326935695 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 6 |
Hb_004463_040 |
0.1342923616 |
- |
- |
- |
| 7 |
Hb_000666_020 |
0.134388414 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 8 |
Hb_001902_180 |
0.1355749091 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 9 |
Hb_003900_050 |
0.136885257 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 10 |
Hb_000189_260 |
0.1407644819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000684_020 |
0.1432576879 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_002890_400 |
0.1436690824 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 13 |
Hb_000522_040 |
0.1439606151 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 14 |
Hb_002837_140 |
0.1463366739 |
- |
- |
hypothetical protein JCGZ_05022 [Jatropha curcas] |
| 15 |
Hb_110218_010 |
0.1466292726 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 16 |
Hb_020581_020 |
0.1470188872 |
- |
- |
hypothetical protein B456_003G065300 [Gossypium raimondii] |
| 17 |
Hb_064398_010 |
0.1474236108 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_011282_070 |
0.1481734423 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_004200_040 |
0.150668473 |
- |
- |
laccase, putative [Ricinus communis] |
| 20 |
Hb_021419_060 |
0.1520531671 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |