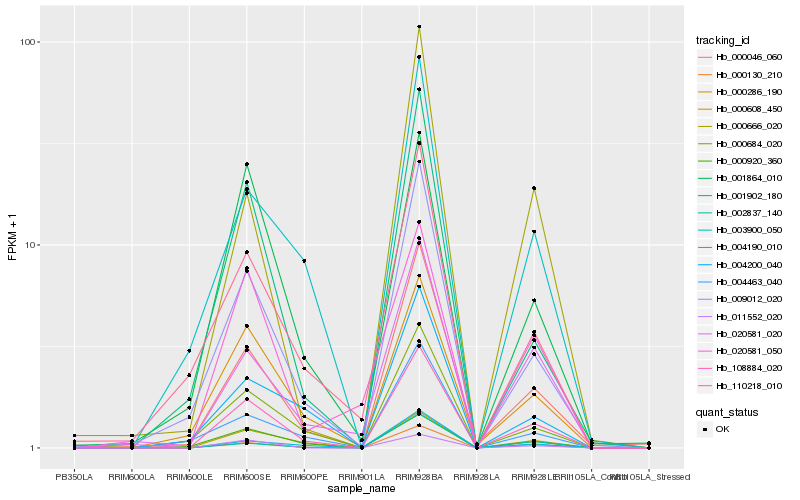
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108884_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 2 |
Hb_000608_450 |
0.1033134311 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 3 |
Hb_000046_060 |
0.1092136964 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 4 |
Hb_009012_020 |
0.1178089886 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 5 |
Hb_000286_190 |
0.1259555339 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 6 |
Hb_004190_010 |
0.1347506807 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000684_020 |
0.1365654258 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_000130_210 |
0.1367228953 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Populus euphratica] |
| 9 |
Hb_020581_020 |
0.1418467808 |
- |
- |
hypothetical protein B456_003G065300 [Gossypium raimondii] |
| 10 |
Hb_003900_050 |
0.1430899431 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 11 |
Hb_000920_360 |
0.1440920817 |
- |
- |
PREDICTED: (R,S)-reticuline 7-O-methyltransferase-like [Jatropha curcas] |
| 12 |
Hb_011552_020 |
0.1458918693 |
- |
- |
hypothetical protein F383_28301 [Gossypium arboreum] |
| 13 |
Hb_001902_180 |
0.1469772733 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 14 |
Hb_004200_040 |
0.1526302931 |
- |
- |
laccase, putative [Ricinus communis] |
| 15 |
Hb_110218_010 |
0.1588161931 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 16 |
Hb_004463_040 |
0.1603750859 |
- |
- |
- |
| 17 |
Hb_001864_010 |
0.160869957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_020581_050 |
0.1644352506 |
- |
- |
- |
| 19 |
Hb_002837_140 |
0.1689590288 |
- |
- |
hypothetical protein JCGZ_05022 [Jatropha curcas] |
| 20 |
Hb_000666_020 |
0.1691220021 |
- |
- |
Major latex protein, putative [Ricinus communis] |