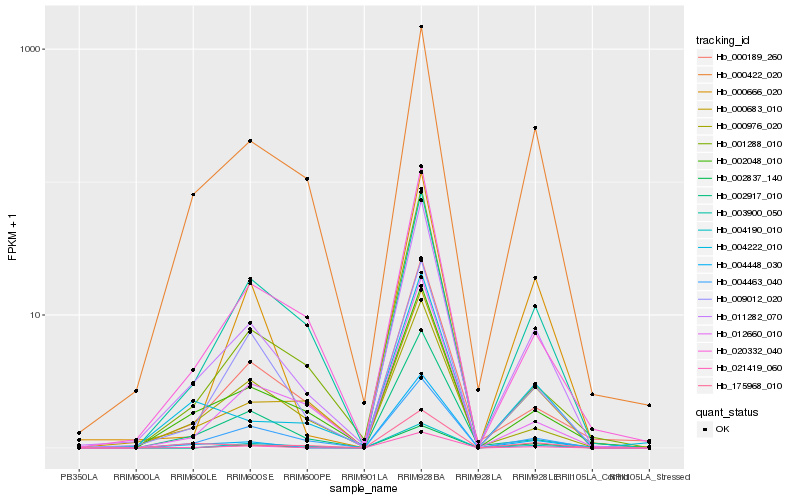
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011282_070 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_021419_060 |
0.0586095267 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 3 |
Hb_004463_040 |
0.1104397685 |
- |
- |
- |
| 4 |
Hb_002048_010 |
0.1117041008 |
- |
- |
unknown [Lotus japonicus] |
| 5 |
Hb_020332_040 |
0.1175080126 |
- |
- |
PREDICTED: alcohol dehydrogenase 1-like [Jatropha curcas] |
| 6 |
Hb_000422_020 |
0.1178957054 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 7 |
Hb_000683_010 |
0.1215514358 |
- |
- |
Interleukin-1 receptor-associated kinase, putative [Ricinus communis] |
| 8 |
Hb_000189_260 |
0.130009784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_175968_010 |
0.1343238923 |
- |
- |
hypothetical protein JCGZ_21281 [Jatropha curcas] |
| 10 |
Hb_002837_140 |
0.1356423646 |
- |
- |
hypothetical protein JCGZ_05022 [Jatropha curcas] |
| 11 |
Hb_002917_010 |
0.1358817567 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 12 |
Hb_009012_020 |
0.1371128959 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 13 |
Hb_000666_020 |
0.1384105551 |
- |
- |
Major latex protein, putative [Ricinus communis] |
| 14 |
Hb_004190_010 |
0.1386986095 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_012660_010 |
0.1413098531 |
- |
- |
hypothetical protein JCGZ_25971 [Jatropha curcas] |
| 16 |
Hb_003900_050 |
0.1424945957 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 17 |
Hb_004222_010 |
0.142823146 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 18 |
Hb_001288_010 |
0.1438158337 |
- |
- |
- |
| 19 |
Hb_000976_020 |
0.1442504858 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 20 |
Hb_004448_030 |
0.1455116806 |
- |
- |
PREDICTED: RING-H2 finger protein ATL40-like [Jatropha curcas] |