| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
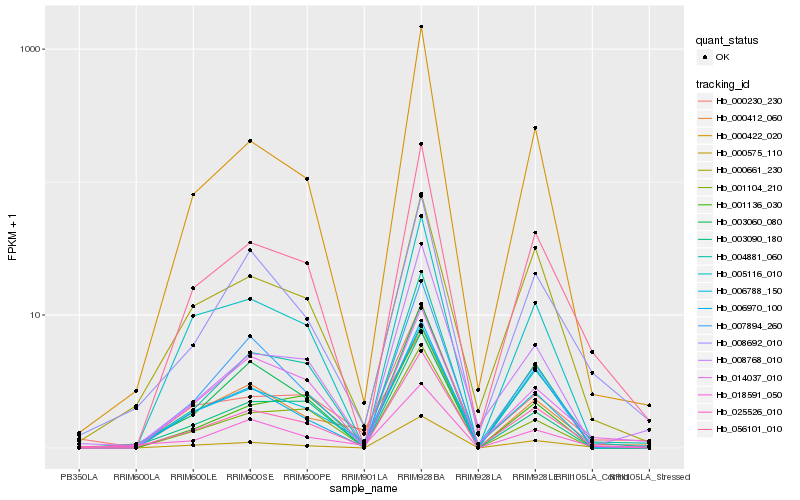
Hb_025526_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 2 |
Hb_006788_150 |
0.0836950897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_056101_010 |
0.0841903665 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |
| 4 |
Hb_000661_230 |
0.0977387807 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000422_020 |
0.1081170208 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_004881_060 |
0.1094183068 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 7 |
Hb_003060_080 |
0.1109800904 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_008692_010 |
0.1139732721 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 9 |
Hb_005116_010 |
0.1179950851 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 10 |
Hb_007894_260 |
0.1185132026 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_003090_180 |
0.1216047003 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 12 |
Hb_008768_010 |
0.1270025141 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000230_230 |
0.1273512992 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 14 |
Hb_018591_050 |
0.129619386 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 15 |
Hb_006970_100 |
0.1304652038 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000575_110 |
0.1340152197 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 17 |
Hb_001104_210 |
0.1348408574 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |
| 18 |
Hb_001136_030 |
0.1385983082 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |
| 19 |
Hb_014037_010 |
0.141706393 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 20 |
Hb_000412_060 |
0.1435785899 |
- |
- |
hypothetical protein JCGZ_18814 [Jatropha curcas] |