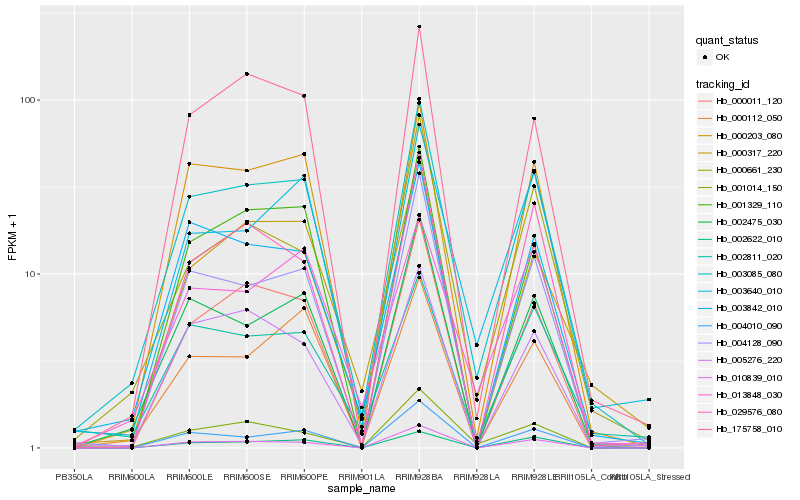
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003085_080 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_004128_090 |
0.0974047692 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 3 |
Hb_002475_030 |
0.098160698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003842_010 |
0.1009282535 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000203_080 |
0.105953006 |
- |
- |
Xylem bark cysteine peptidase 3 isoform 1 [Theobroma cacao] |
| 6 |
Hb_010839_010 |
0.1139218673 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_001014_150 |
0.1147508604 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_003640_010 |
0.1186432793 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 9 |
Hb_000011_120 |
0.1187315615 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 10 |
Hb_002811_020 |
0.1231688755 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 11 |
Hb_004010_090 |
0.1232217596 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_175758_010 |
0.1243374529 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_005276_220 |
0.1243466047 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 14 |
Hb_000661_230 |
0.1270376332 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_013848_030 |
0.1286514949 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 16 |
Hb_029576_080 |
0.1286709488 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 17 |
Hb_000317_220 |
0.1291115605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001329_110 |
0.1295348897 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 19 |
Hb_002622_010 |
0.1302065221 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 20 |
Hb_000112_050 |
0.1304738298 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |