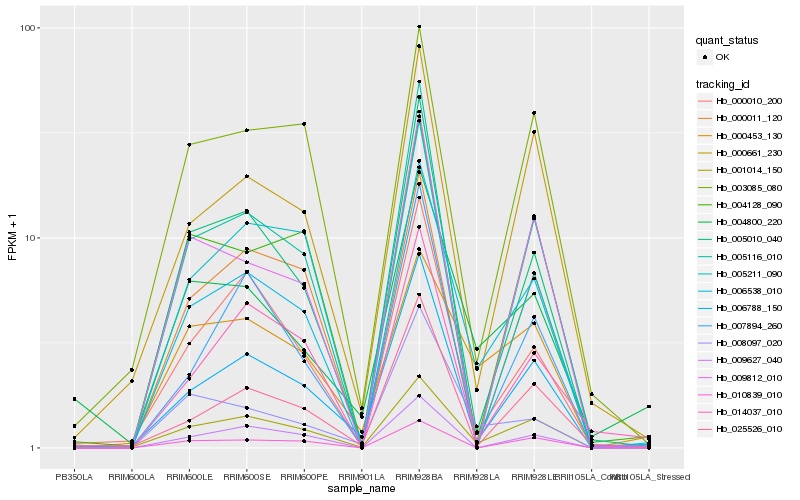
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006538_010 |
0.0 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 2 |
Hb_001014_150 |
0.0987548029 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_006788_150 |
0.1247361826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005010_040 |
0.1364576963 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 5 |
Hb_005116_010 |
0.1368498435 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 6 |
Hb_009627_040 |
0.1465229869 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 7 |
Hb_000661_230 |
0.1470414847 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000010_200 |
0.1476988996 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_010839_010 |
0.1504165634 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 10 |
Hb_025526_010 |
0.1568423268 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 11 |
Hb_004128_090 |
0.1573894926 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 12 |
Hb_000453_130 |
0.1593944127 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_008097_020 |
0.1596219318 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 14 |
Hb_009812_010 |
0.1602811301 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_014037_010 |
0.1607388961 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 16 |
Hb_003085_080 |
0.1613244076 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_007894_260 |
0.1631331439 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_004800_220 |
0.1638359176 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 19 |
Hb_000011_120 |
0.1638570733 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 20 |
Hb_005211_090 |
0.1638777986 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |