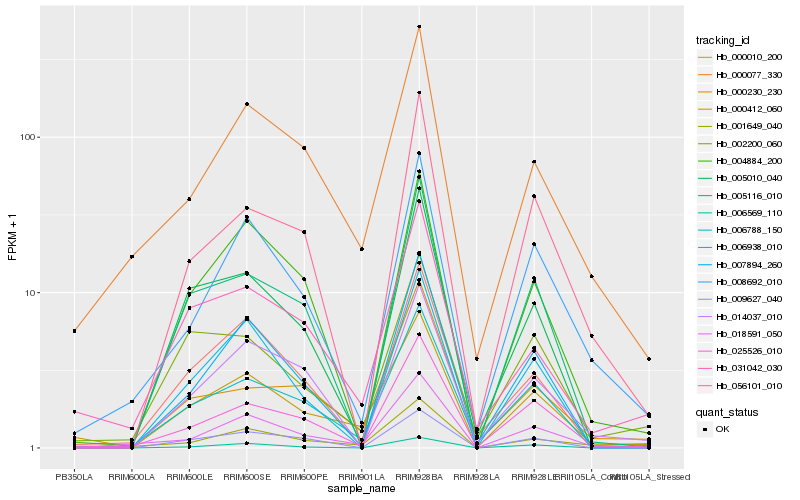
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000412_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_18814 [Jatropha curcas] |
| 2 |
Hb_006788_150 |
0.1172573684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_018591_050 |
0.1345574509 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 4 |
Hb_025526_010 |
0.1435785899 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 5 |
Hb_007894_260 |
0.1501999696 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_005116_010 |
0.1510527501 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 7 |
Hb_000077_330 |
0.1518417951 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 8 |
Hb_004884_200 |
0.1538992528 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002200_060 |
0.1554590822 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8 [Jatropha curcas] |
| 10 |
Hb_008692_010 |
0.1565835562 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 11 |
Hb_005010_040 |
0.1567063425 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 12 |
Hb_006938_010 |
0.1571658458 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 13 |
Hb_031042_030 |
0.1612476549 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_009627_040 |
0.1612594356 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 15 |
Hb_001649_040 |
0.1618537108 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 16 |
Hb_000010_200 |
0.1626242315 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_014037_010 |
0.1632124593 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 18 |
Hb_000230_230 |
0.1634870689 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 19 |
Hb_006569_110 |
0.1637540691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_056101_010 |
0.1647944773 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |