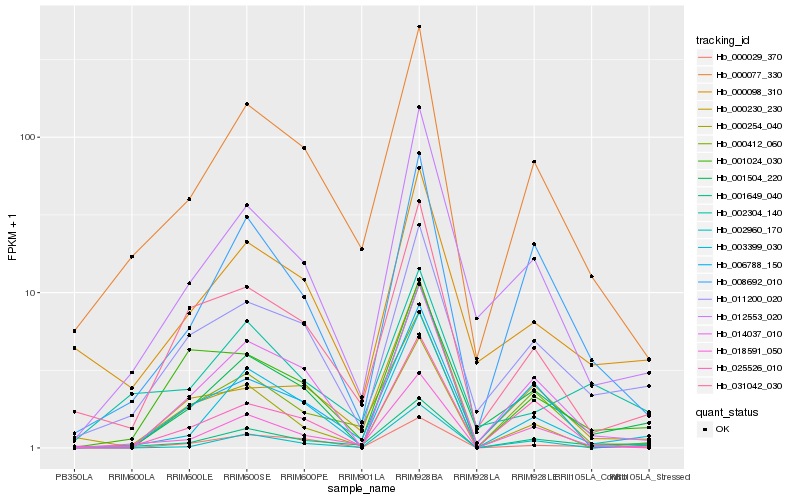
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001649_040 |
0.0 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 2 |
Hb_000077_330 |
0.1370743574 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 3 |
Hb_003399_030 |
0.1453978036 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 4 |
Hb_011200_020 |
0.1512049664 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 5 |
Hb_018591_050 |
0.1519301764 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 6 |
Hb_001504_220 |
0.1609275424 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_008692_010 |
0.1609857848 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 8 |
Hb_000412_060 |
0.1618537108 |
- |
- |
hypothetical protein JCGZ_18814 [Jatropha curcas] |
| 9 |
Hb_012553_020 |
0.1704805168 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 10 |
Hb_002960_170 |
0.1807102008 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 11 |
Hb_031042_030 |
0.1832413838 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000098_310 |
0.1841193131 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 13 |
Hb_000029_370 |
0.1854827958 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 14 |
Hb_002304_140 |
0.1880065727 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 15 |
Hb_001024_030 |
0.1894678615 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_014037_010 |
0.192607063 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 17 |
Hb_025526_010 |
0.1942871399 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 18 |
Hb_000230_230 |
0.1948582388 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 19 |
Hb_000254_040 |
0.1958505481 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 20 |
Hb_006788_150 |
0.1990485673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |