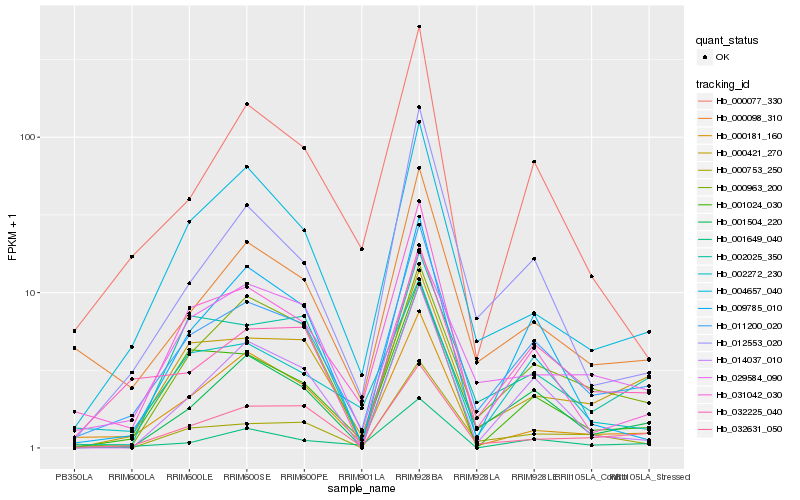
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011200_020 |
0.0 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 2 |
Hb_000098_310 |
0.1226424246 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 3 |
Hb_001504_220 |
0.1261511971 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_032225_040 |
0.1283162328 |
- |
- |
- |
| 5 |
Hb_031042_030 |
0.1367894092 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_032631_050 |
0.1370904711 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 7 |
Hb_000421_270 |
0.1372891779 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 8 |
Hb_001024_030 |
0.1444522433 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002272_230 |
0.1446657531 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 10 |
Hb_000963_200 |
0.1478760967 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 11 |
Hb_004657_040 |
0.1483353382 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 12 |
Hb_000077_330 |
0.1502152906 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 13 |
Hb_001649_040 |
0.1512049664 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 14 |
Hb_002025_350 |
0.1514496749 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 15 |
Hb_012553_020 |
0.155284876 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 16 |
Hb_000181_160 |
0.1579560273 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000753_250 |
0.1601340194 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 18 |
Hb_014037_010 |
0.1601640372 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 19 |
Hb_029584_090 |
0.1649224735 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 20 |
Hb_009785_010 |
0.1652552264 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |