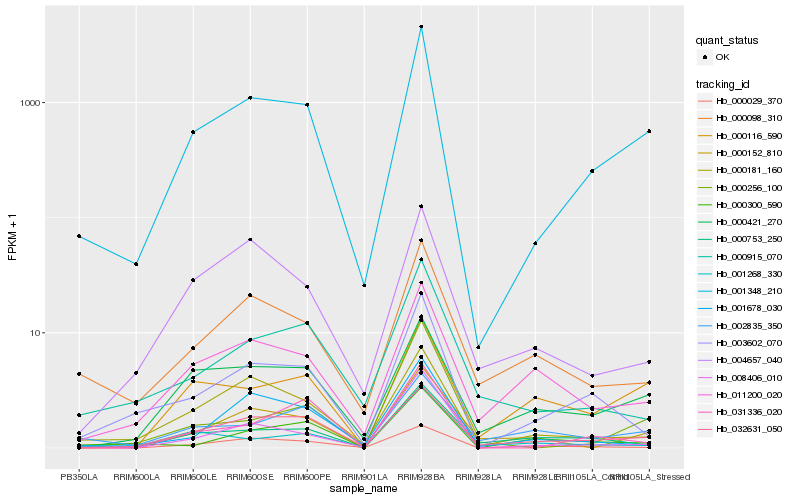
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_210 |
0.0 |
- |
- |
defensin protein precursor [Capsicum annuum] |
| 2 |
Hb_032631_050 |
0.1355450588 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 3 |
Hb_000421_270 |
0.1740085027 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 4 |
Hb_000181_160 |
0.1744911958 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003602_070 |
0.1853043943 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 6 |
Hb_000116_590 |
0.1964923776 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 7 |
Hb_000098_310 |
0.1981394193 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 8 |
Hb_011200_020 |
0.1986871102 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 9 |
Hb_000029_370 |
0.2002260236 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 10 |
Hb_008406_010 |
0.2023025935 |
- |
- |
PREDICTED: tyrosine aminotransferase-like [Populus euphratica] |
| 11 |
Hb_000152_810 |
0.205529383 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 12 |
Hb_001678_030 |
0.2068705275 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 13 |
Hb_002835_350 |
0.207404028 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 14 |
Hb_000915_070 |
0.2093196373 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_004657_040 |
0.2138407665 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 16 |
Hb_031336_020 |
0.2152618456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000753_250 |
0.2157271997 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 18 |
Hb_000256_100 |
0.2163379763 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 19 |
Hb_001268_330 |
0.2174672734 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_000300_590 |
0.2193668769 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |