| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
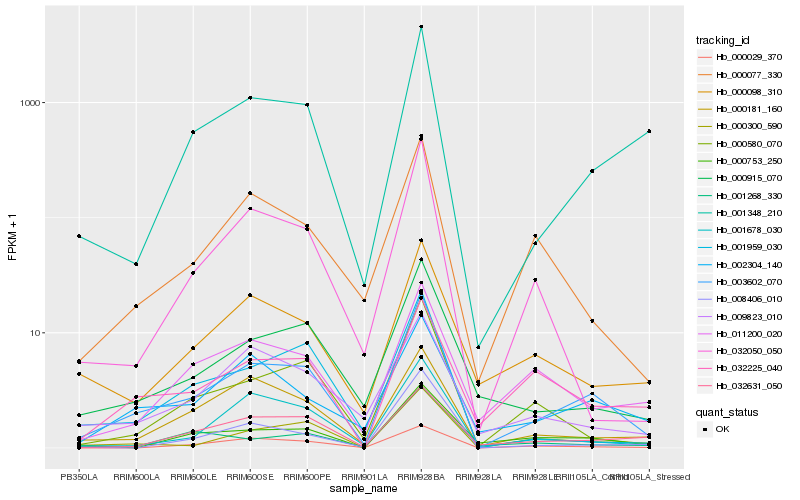
Hb_003602_070 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 2 |
Hb_000300_590 |
0.1635104296 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 3 |
Hb_000915_070 |
0.1730627906 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000580_070 |
0.1754993301 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 5 |
Hb_001678_030 |
0.1774762572 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_000181_160 |
0.1830368578 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002304_140 |
0.184217771 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_000029_370 |
0.1845701929 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 9 |
Hb_001348_210 |
0.1853043943 |
- |
- |
defensin protein precursor [Capsicum annuum] |
| 10 |
Hb_000077_330 |
0.1906963144 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2-like [Jatropha curcas] |
| 11 |
Hb_001959_030 |
0.1919442159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000753_250 |
0.191998444 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 13 |
Hb_000098_310 |
0.1944179173 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 14 |
Hb_032050_050 |
0.1958454668 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 15 |
Hb_032631_050 |
0.1964111789 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 16 |
Hb_011200_020 |
0.1985412969 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 17 |
Hb_032225_040 |
0.1992291265 |
- |
- |
- |
| 18 |
Hb_008406_010 |
0.2025257275 |
- |
- |
PREDICTED: tyrosine aminotransferase-like [Populus euphratica] |
| 19 |
Hb_009823_010 |
0.2028070631 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 20 |
Hb_001268_330 |
0.2048151769 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |