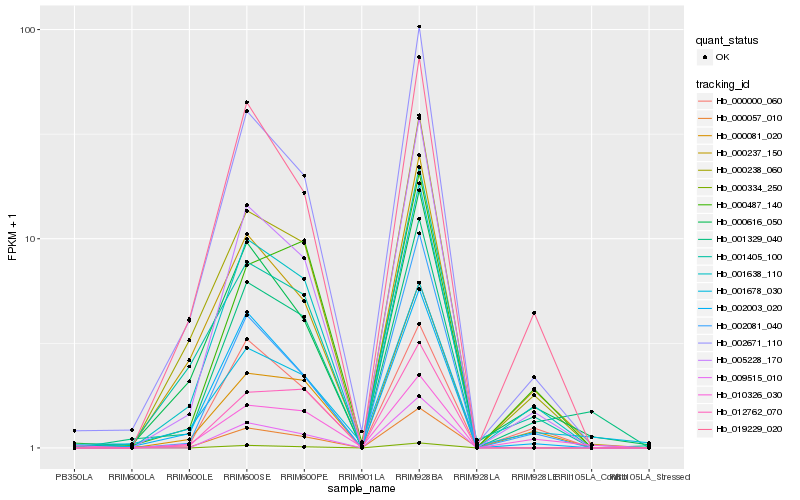
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_040 |
0.0 |
- |
- |
hypothetical protein RCOM_1345340 [Ricinus communis] |
| 2 |
Hb_001678_030 |
0.1254092233 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 3 |
Hb_001638_110 |
0.1276556374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005228_170 |
0.1368820167 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 5 |
Hb_010326_030 |
0.1394531702 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 6 |
Hb_002671_110 |
0.1400184902 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 7 |
Hb_000616_050 |
0.1417756443 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 8 |
Hb_019229_020 |
0.1519608152 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000238_060 |
0.1573498778 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 10 |
Hb_000237_150 |
0.1584831726 |
- |
- |
Epoxide hydrolase 2 [Morus notabilis] |
| 11 |
Hb_000081_020 |
0.1612525278 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 1.1-like [Camelina sativa] |
| 12 |
Hb_002003_020 |
0.1629792765 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 13 |
Hb_000487_140 |
0.1638908771 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000057_010 |
0.1649849685 |
- |
- |
PREDICTED: probable protein phosphatase 2C 59 [Jatropha curcas] |
| 15 |
Hb_002081_040 |
0.1649979859 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 16 |
Hb_009515_010 |
0.1675878102 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 17 |
Hb_000334_250 |
0.1680609496 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 18 |
Hb_001405_100 |
0.1684136089 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 19 |
Hb_012762_070 |
0.1685713282 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 20 |
Hb_000000_060 |
0.1687409787 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |