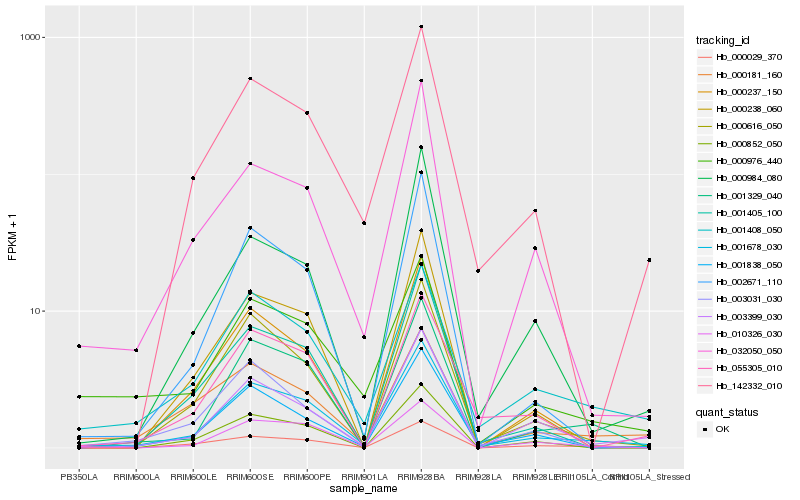
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001678_030 |
0.0 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 2 |
Hb_000616_050 |
0.1183566309 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 3 |
Hb_001408_050 |
0.1190309169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000029_370 |
0.1247120273 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 5 |
Hb_001329_040 |
0.1254092233 |
- |
- |
hypothetical protein RCOM_1345340 [Ricinus communis] |
| 6 |
Hb_001838_050 |
0.129183097 |
- |
- |
hypothetical protein B456_007G218500 [Gossypium raimondii] |
| 7 |
Hb_032050_050 |
0.1321396823 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 8 |
Hb_002671_110 |
0.1325632842 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 9 |
Hb_001405_100 |
0.1326068339 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 10 |
Hb_003399_030 |
0.1326179889 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 11 |
Hb_142332_010 |
0.1329529612 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 12 |
Hb_010326_030 |
0.1334837268 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 13 |
Hb_000237_150 |
0.1368858566 |
- |
- |
Epoxide hydrolase 2 [Morus notabilis] |
| 14 |
Hb_000181_160 |
0.1401822557 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000852_050 |
0.1407208297 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 16 |
Hb_000976_440 |
0.1432402715 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_003031_030 |
0.1454093737 |
- |
- |
hypothetical protein JCGZ_21288 [Jatropha curcas] |
| 18 |
Hb_000238_060 |
0.1467624945 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 19 |
Hb_000984_080 |
0.1480013107 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 20 |
Hb_055305_010 |
0.1485102938 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |