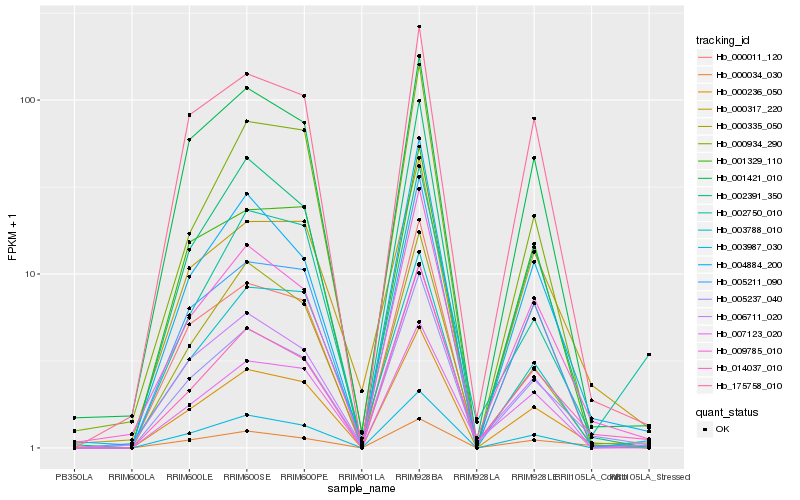
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_050 |
0.0 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 2 |
Hb_005237_040 |
0.0778370058 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 3 |
Hb_009785_010 |
0.0914130366 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |
| 4 |
Hb_014037_010 |
0.0947641032 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 5 |
Hb_004884_200 |
0.098559155 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005211_090 |
0.1038364328 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002391_350 |
0.1040234331 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 8 |
Hb_000317_220 |
0.1071500798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006711_020 |
0.1115208762 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 10 |
Hb_000934_290 |
0.1120066392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_175758_010 |
0.1134993309 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000034_030 |
0.1147940077 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_001421_010 |
0.1156435024 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001329_110 |
0.1158983742 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 15 |
Hb_003987_030 |
0.116272828 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 16 |
Hb_000011_120 |
0.117073927 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 17 |
Hb_000335_050 |
0.1206104597 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007123_020 |
0.1214102141 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 19 |
Hb_003788_010 |
0.121548851 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 20 |
Hb_002750_010 |
0.1229421654 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |