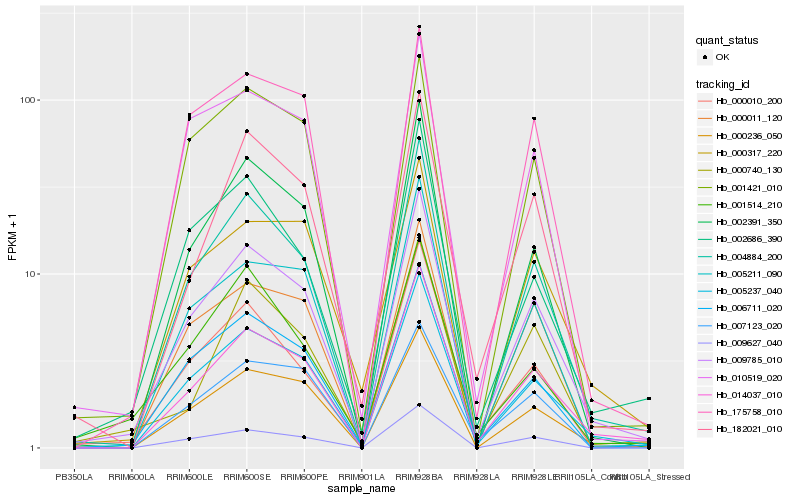
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009785_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |
| 2 |
Hb_005237_040 |
0.0606395581 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 3 |
Hb_004884_200 |
0.0612844097 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_014037_010 |
0.0758483555 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 5 |
Hb_006711_020 |
0.0861072252 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 6 |
Hb_000236_050 |
0.0914130366 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_002391_350 |
0.0947356393 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 8 |
Hb_009627_040 |
0.1029890255 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 9 |
Hb_000010_200 |
0.1038125962 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_002686_390 |
0.1087847997 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 11 |
Hb_005211_090 |
0.1114497734 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_182021_010 |
0.1115707352 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 13 |
Hb_000011_120 |
0.1136571591 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 14 |
Hb_010519_020 |
0.1142731724 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 15 |
Hb_175758_010 |
0.1172169653 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000317_220 |
0.119269885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001421_010 |
0.1228379752 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007123_020 |
0.1257412056 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 19 |
Hb_000740_130 |
0.1275441612 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 20 |
Hb_001514_210 |
0.1283668194 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |