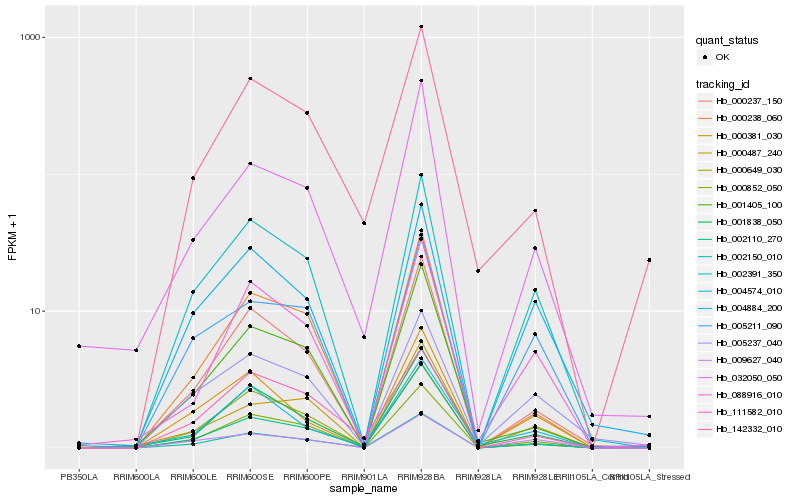
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s10240g [Populus trichocarpa] |
| 2 |
Hb_002391_350 |
0.0913871994 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 3 |
Hb_000237_150 |
0.09139611 |
- |
- |
Epoxide hydrolase 2 [Morus notabilis] |
| 4 |
Hb_000852_050 |
0.0914057941 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 5 |
Hb_032050_050 |
0.1075722893 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 6 |
Hb_001838_050 |
0.1081411738 |
- |
- |
hypothetical protein B456_007G218500 [Gossypium raimondii] |
| 7 |
Hb_088916_010 |
0.118174899 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 8 |
Hb_000238_060 |
0.1205193083 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 9 |
Hb_002110_270 |
0.1220871163 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 10 |
Hb_000649_030 |
0.1227258043 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 11 |
Hb_000381_030 |
0.1227332771 |
- |
- |
PREDICTED: aquaporin SIP1-1-like [Jatropha curcas] |
| 12 |
Hb_001405_100 |
0.1244622295 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 13 |
Hb_142332_010 |
0.1252847541 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 14 |
Hb_004884_200 |
0.125876745 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_111582_010 |
0.1268473509 |
- |
- |
hypothetical protein POPTR_0303s002201g, partial [Populus trichocarpa] |
| 16 |
Hb_005211_090 |
0.1279795099 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_009627_040 |
0.1305749936 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 18 |
Hb_005237_040 |
0.1330700073 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 19 |
Hb_000487_240 |
0.1331512341 |
- |
- |
PREDICTED: CYSTM1 family protein B [Cucumis melo] |
| 20 |
Hb_004574_010 |
0.1332844045 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |