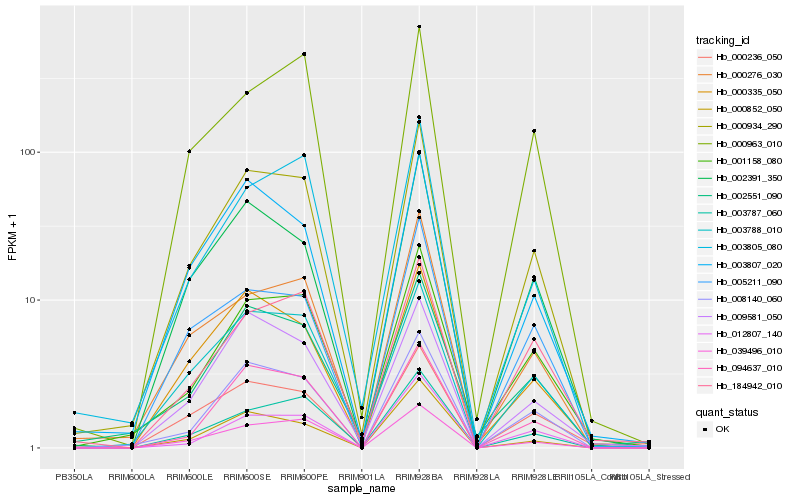
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000934_290 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008140_060 |
0.0729957719 |
- |
- |
Cytochrome P450, putative [Theobroma cacao] |
| 3 |
Hb_003788_010 |
0.0808764571 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 4 |
Hb_039496_010 |
0.0829446487 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 5 |
Hb_001158_080 |
0.0862700874 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 6 |
Hb_003787_060 |
0.090332263 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 7 |
Hb_094637_010 |
0.0937370851 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |
| 8 |
Hb_002391_350 |
0.097254191 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 9 |
Hb_003807_020 |
0.1003891062 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 10 |
Hb_000335_050 |
0.102031272 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002551_090 |
0.1020868898 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 12 |
Hb_003805_080 |
0.1063058711 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 13 |
Hb_000276_030 |
0.1075651939 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_000852_050 |
0.1090028842 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 15 |
Hb_009581_050 |
0.1090617625 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |
| 16 |
Hb_000236_050 |
0.1120066392 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 17 |
Hb_184942_010 |
0.1144791013 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 18 |
Hb_000963_010 |
0.115873339 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 19 |
Hb_005211_090 |
0.1159309065 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_012807_140 |
0.1175420883 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |