| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
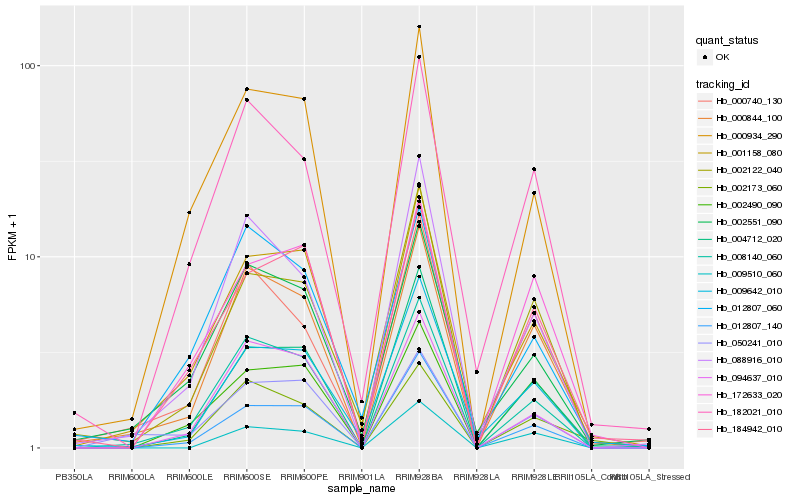
Hb_000844_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_04351 [Jatropha curcas] |
| 2 |
Hb_008140_060 |
0.0847629465 |
- |
- |
Cytochrome P450, putative [Theobroma cacao] |
| 3 |
Hb_000740_130 |
0.1006539463 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 4 |
Hb_182021_010 |
0.1103720992 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 5 |
Hb_001158_080 |
0.1136406053 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 6 |
Hb_002173_060 |
0.1146790377 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 7 |
Hb_094637_010 |
0.1230352387 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |
| 8 |
Hb_088916_010 |
0.1231513185 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 9 |
Hb_002551_090 |
0.1245711596 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 10 |
Hb_002122_040 |
0.1256282585 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_000934_290 |
0.1273669468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_050241_010 |
0.1304539945 |
- |
- |
PREDICTED: uncharacterized protein LOC105129233 isoform X1 [Populus euphratica] |
| 13 |
Hb_002490_090 |
0.1307919495 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 14 |
Hb_004712_020 |
0.1331356245 |
- |
- |
hypothetical protein JCGZ_00782 [Jatropha curcas] |
| 15 |
Hb_009642_010 |
0.1354924749 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 16 |
Hb_012807_140 |
0.136460695 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 17 |
Hb_172633_020 |
0.1367141313 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 [Populus euphratica] |
| 18 |
Hb_009510_060 |
0.1370736612 |
- |
- |
- |
| 19 |
Hb_184942_010 |
0.1411357719 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 20 |
Hb_012807_060 |
0.1412607758 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |