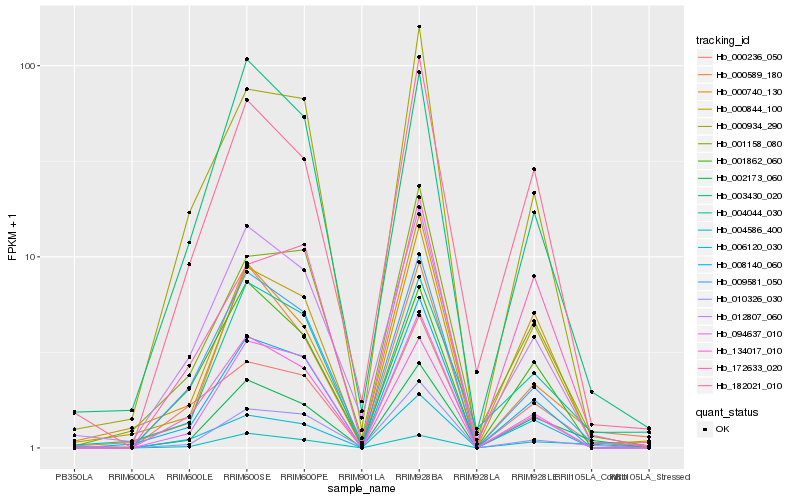
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002173_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 2 |
Hb_000844_100 |
0.1146790377 |
- |
- |
hypothetical protein JCGZ_04351 [Jatropha curcas] |
| 3 |
Hb_182021_010 |
0.1320750371 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 4 |
Hb_008140_060 |
0.1339183023 |
- |
- |
Cytochrome P450, putative [Theobroma cacao] |
| 5 |
Hb_000589_180 |
0.1359596219 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 6 |
Hb_009581_050 |
0.1403577422 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |
| 7 |
Hb_094637_010 |
0.1409480251 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |
| 8 |
Hb_004044_030 |
0.1506214241 |
- |
- |
PREDICTED: uncharacterized protein LOC105639335 [Jatropha curcas] |
| 9 |
Hb_012807_060 |
0.1525826549 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_004586_400 |
0.1525910364 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_003430_020 |
0.1541225455 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_001862_060 |
0.1544993151 |
- |
- |
PREDICTED: uncharacterized protein LOC105639802 isoform X1 [Jatropha curcas] |
| 13 |
Hb_010326_030 |
0.1580515527 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 14 |
Hb_000740_130 |
0.1581306675 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 15 |
Hb_000236_050 |
0.1590717585 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 16 |
Hb_000934_290 |
0.1594780554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001158_080 |
0.1602439664 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 18 |
Hb_172633_020 |
0.1606503734 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 [Populus euphratica] |
| 19 |
Hb_006120_030 |
0.1621151318 |
- |
- |
- |
| 20 |
Hb_134017_010 |
0.1625734581 |
- |
- |
kinase, putative [Ricinus communis] |