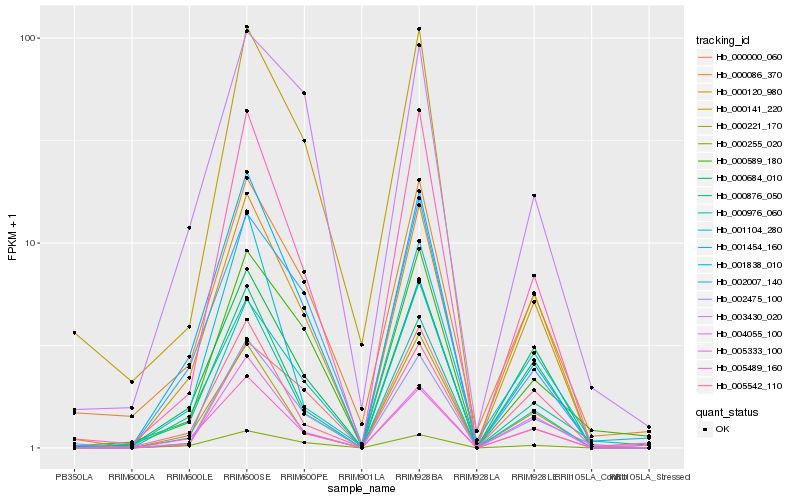
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002475_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 2 |
Hb_005489_160 |
0.1115766026 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 3 |
Hb_000120_980 |
0.1193265619 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 4 |
Hb_000589_180 |
0.1246567273 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 5 |
Hb_005333_100 |
0.1302091553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001838_010 |
0.1374073319 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 7 |
Hb_000086_370 |
0.1377154755 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 8 |
Hb_000976_060 |
0.1379666386 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000684_010 |
0.1426128335 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_001104_280 |
0.1434089802 |
- |
- |
PREDICTED: UPF0481 protein At3g47200 [Jatropha curcas] |
| 11 |
Hb_004055_100 |
0.1448083165 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 12 |
Hb_000255_020 |
0.1518509676 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 13 |
Hb_005542_110 |
0.1560743509 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 14 |
Hb_000221_170 |
0.1602178374 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 15 |
Hb_000000_060 |
0.1623584914 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 16 |
Hb_001454_160 |
0.1634412175 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000876_050 |
0.1644491053 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 18 |
Hb_002007_140 |
0.1651110631 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 19 |
Hb_003430_020 |
0.167448806 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_000141_220 |
0.1675991871 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |