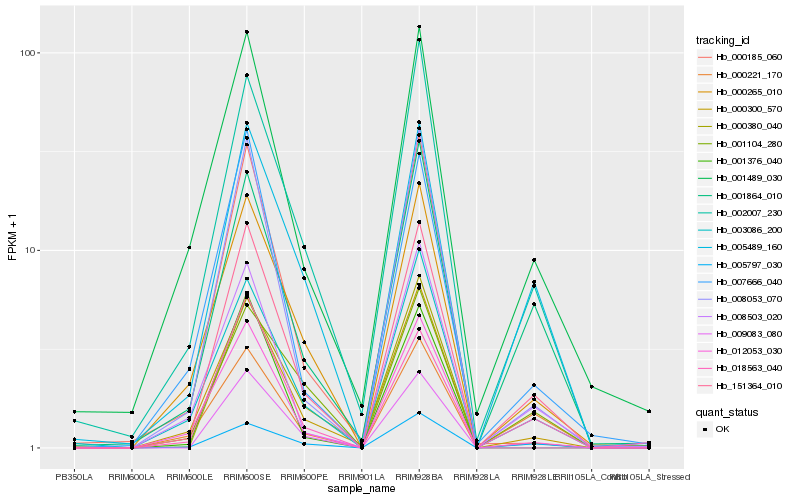
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_151364_010 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 2 |
Hb_005489_160 |
0.0969369538 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 3 |
Hb_008053_070 |
0.1039165545 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X4 [Jatropha curcas] |
| 4 |
Hb_000300_570 |
0.1049179535 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 5 |
Hb_012053_030 |
0.105288162 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 6 |
Hb_005797_030 |
0.105827018 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 7 |
Hb_002007_230 |
0.106815978 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 8 |
Hb_000265_010 |
0.1075128201 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_001864_010 |
0.1143049178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008503_020 |
0.1143754797 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_003086_200 |
0.1147498292 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_007666_040 |
0.1234539618 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000380_040 |
0.1237631512 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 14 |
Hb_001489_030 |
0.1263132355 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 15 |
Hb_018563_040 |
0.1267280924 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000185_060 |
0.130622232 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001104_280 |
0.1328245779 |
- |
- |
PREDICTED: UPF0481 protein At3g47200 [Jatropha curcas] |
| 18 |
Hb_009083_080 |
0.1374735936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000221_170 |
0.14098836 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 20 |
Hb_001376_040 |
0.1422405533 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |