| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
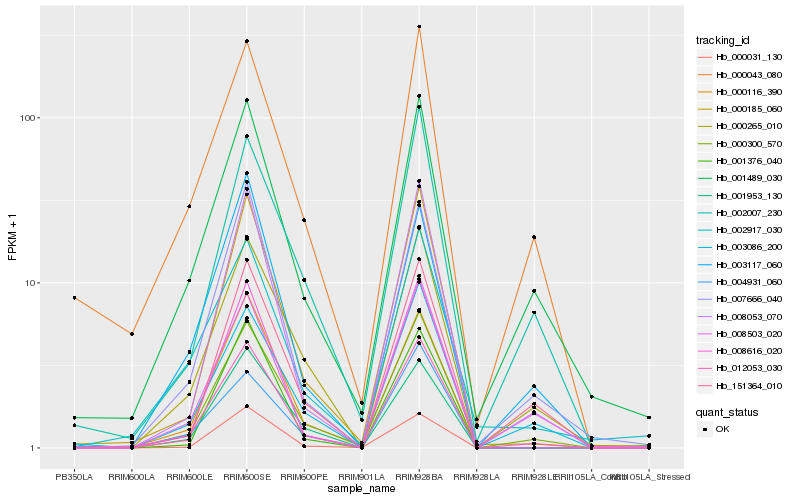
Hb_012053_030 |
0.0 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 2 |
Hb_000300_570 |
0.0649527062 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 3 |
Hb_000265_010 |
0.0819878099 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_008053_070 |
0.0830298594 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X4 [Jatropha curcas] |
| 5 |
Hb_000185_060 |
0.0927241719 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003086_200 |
0.0974757078 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 7 |
Hb_007666_040 |
0.0999679715 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_008616_020 |
0.1012891144 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_008503_020 |
0.1026319782 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 10 |
Hb_001489_030 |
0.104467383 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 11 |
Hb_151364_010 |
0.105288162 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 12 |
Hb_000116_390 |
0.1097877341 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 13 |
Hb_001953_130 |
0.1109710458 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 14 |
Hb_000043_080 |
0.1184212957 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 15 |
Hb_001376_040 |
0.1191234765 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 16 |
Hb_003117_060 |
0.1212071695 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 17 |
Hb_004931_060 |
0.1223684546 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 18 |
Hb_002007_230 |
0.1226228555 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 19 |
Hb_000031_130 |
0.123366099 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 20 |
Hb_002917_030 |
0.1245290632 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Setaria italica] |