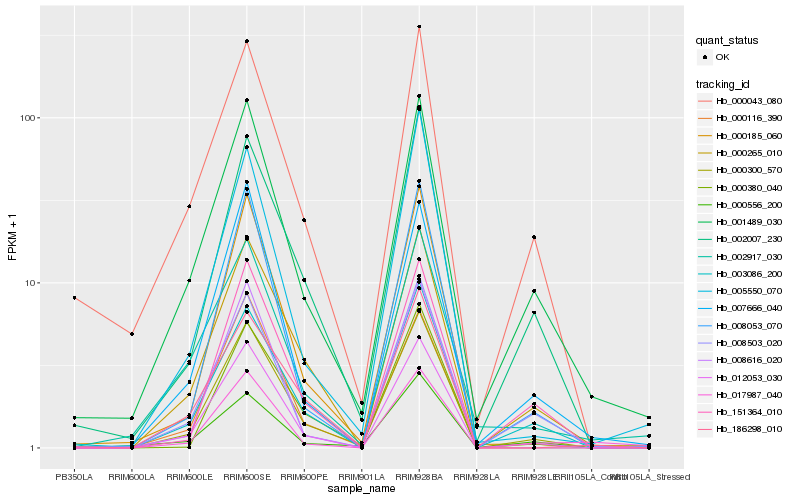
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_570 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 2 |
Hb_012053_030 |
0.0649527062 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 3 |
Hb_000265_010 |
0.0736400381 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_003086_200 |
0.092693543 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 5 |
Hb_002007_230 |
0.0936278868 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 6 |
Hb_001489_030 |
0.0984597303 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 7 |
Hb_000185_060 |
0.0985979241 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008053_070 |
0.103668394 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X4 [Jatropha curcas] |
| 9 |
Hb_151364_010 |
0.1049179535 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 10 |
Hb_008503_020 |
0.1058182827 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_000380_040 |
0.107619507 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 12 |
Hb_000043_080 |
0.1200168621 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 13 |
Hb_005550_070 |
0.1209975594 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 14 |
Hb_000556_200 |
0.1235378793 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 22 [Jatropha curcas] |
| 15 |
Hb_000116_390 |
0.1245501363 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 16 |
Hb_002917_030 |
0.1259887382 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Setaria italica] |
| 17 |
Hb_186298_010 |
0.1283435538 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_017987_040 |
0.1288223486 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 19 |
Hb_007666_040 |
0.1289978163 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_008616_020 |
0.1296249803 |
- |
- |
unnamed protein product [Vitis vinifera] |