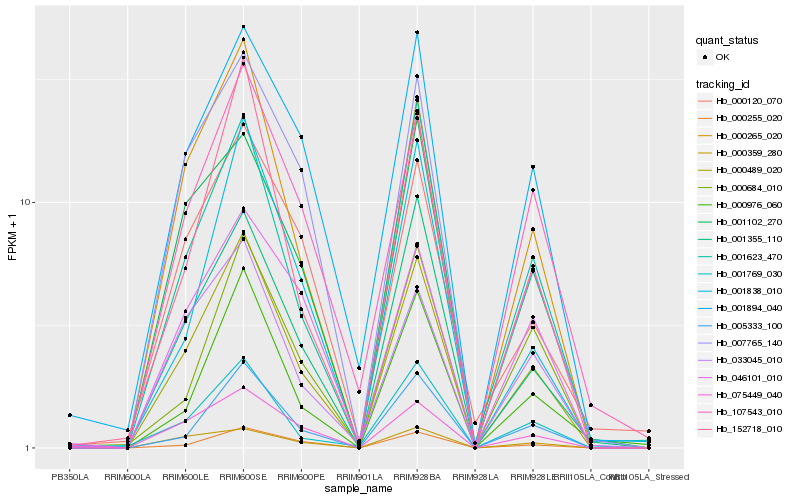
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000489_020 |
0.0 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 2 |
Hb_000265_020 |
0.081672672 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 3 |
Hb_001355_110 |
0.1011267398 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005333_100 |
0.1030305577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001623_470 |
0.1041972288 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 6 |
Hb_152718_010 |
0.1059052468 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 7 |
Hb_001769_030 |
0.1098087546 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 8 |
Hb_000255_020 |
0.1102678084 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 9 |
Hb_033045_010 |
0.1166073925 |
- |
- |
copine, putative [Ricinus communis] |
| 10 |
Hb_001102_270 |
0.1167124542 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 11 |
Hb_007765_140 |
0.1169456743 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 12 |
Hb_001894_040 |
0.1174060789 |
- |
- |
PREDICTED: carbonic anhydrase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_075449_040 |
0.1181483046 |
- |
- |
PREDICTED: meiotic recombination protein SPO11-1 [Jatropha curcas] |
| 14 |
Hb_107543_010 |
0.1216598322 |
- |
- |
Cysteine-rich RLK 10, putative [Theobroma cacao] |
| 15 |
Hb_000976_060 |
0.1248679413 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000359_280 |
0.1317402401 |
- |
- |
hypothetical protein OsI_15300 [Oryza sativa Indica Group] |
| 17 |
Hb_001838_010 |
0.1323671061 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 18 |
Hb_046101_010 |
0.1366902172 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 19 |
Hb_000684_010 |
0.1372977462 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000120_070 |
0.1385517267 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |